Abstract
Free full text

Structural and Functional Analysis of Domains of the Progesterone Receptor
Abstract
Steroid hormone receptors are multi-domain proteins composed of conserved well-structured regions, such as ligand (LBD) and DNA binding domains (DBD), plus other naturally unstructured regions including the amino-terminal domain (NTD) and the hinge region between the LBD and DBD. The hinge is more than just a flexible region between the DBD and LBD and is capable of binding co-regulatory proteins and the minor groove of DNA flanking hormone response elements. Because the hinge can directly participate in DNA binding it has also been termed the carboxyl terminal extension (CTE) of the DNA binding domain. The CTE and NTD are dynamic regions of the receptor that can adopt multiple conformations depending on the environment of interacting proteins and DNA. Both regions have important regulatory roles for multiple receptor functions that are related to the ability of the CTE and NTD to form multiple active conformations. This review focuses on studies of the CTE and NTD of progesterone receptor (PR), as well as related work with other steroid/nuclear receptors.
1. Introduction
Progesterone receptor (PR) mediates the transcriptional effects of the steroid hormone progesterone, which plays a key role in development, differentiation, and maintenance of female reproductive tissues (Li and O’Malley, 2003). PR is expressed as two isoforms, PR-A and PR-B, which arise from the same gene by utilization of two promoters (Kastner et al., 1990; Li and O’Malley, 2003). The two receptors are identical in the C-terminal ligand binding domain (LBD) and DNA binding domain (DBD) and most of the amino-terminal domain (NTD) except for an extension of the NTD unique to PR-B (Figure 1). PR has two transcriptional activation functions (AFs) within the broader domains, AF1 in the NTD and AF2 within the LBD. Depending on the cell and target gene context, AF1 and AF2 can function independently or they can synergize through an intra-molecular interaction between the N- and C-termini (Dong et al., 2004; Meyer et al., 1990; Meyer et al., 1992; Takimoto et al., 2003; Tetel et al., 1999; Tung et al., 2006). Highly conserved AF2 binds short LXXLL amphipathic helix motifs of co-activators including the p160 family steroid receptor co-activators (SRCs), RIP140 and DRIP/TRAPS (Beato and Klug, 2000; Li and O’Malley, 2003; Lonard and O’Malley B, 2007). The unique amino acids in PR-B NTD contain an additional activation function (AF3), that operates by synergizing with AF1 and AF2 (Takimoto et al., 2003; Tung et al., 2006). For most gene targets PR-B is a stronger transcriptional activator than PR-A, while PR-A can act to suppress PR-B and other nuclear receptors (Tung et al., 1993; Vegeto et al., 1993). Functional differences between the two isoforms are evident from knockout studies in mice, which showed that PR-A null mice have normal mammary gland development while PR-B null mice displayed reduced mammary gland development, indicating the importance of PR-B, but not PR-A, in the mouse mammary gland. In contrast, PR-A has been shown to be critical in the uterus and the ovary (Fernandez-Valdivia et al., 2005). Microarray analysis of breast cancer cells has shown that 65 of 94 PR regulated genes were regulated by PR-B alone, 25 were regulated by both isoforms, and only four were regulated by PR-A alone (Richer et al., 2002). PR-A and PR-B can act as both homodimers and heterodimers and are co-expressed at equivalent levels in the same cell in normal human epithelial cells. In breast cancer, the equivalent expression of PR-A and PR-B is often disrupted resulting in a predominance of one form, usually PR-A (Mote et al., 1999; Mote et al., 2002). PR can also mediate non-genomic responses to progesterone through its ability to recruit and activate c-Src tyrosine kinase. A polyproline motif (aa 421- PPPPLPPR-428) in the NTD of PR (Figure 1) binds directly with the SH3 domain of c-Src resulting in the activation of Src dependent phosphorylation signaling cascades in mammalian cells (Boonyaratanakornkit et al., 2001). These include the proliferative Ras/Raf/MEK/MAK kinase and JAK-1/-2/Stat3 signaling pathways and the PI3K/Akt cell survival pathway (Ballare et al., 2003; Boonyaratanakornkit et al., 2007; Proietti et al., 2005; Saitoh et al., 2005; Skildum et al., 2005).
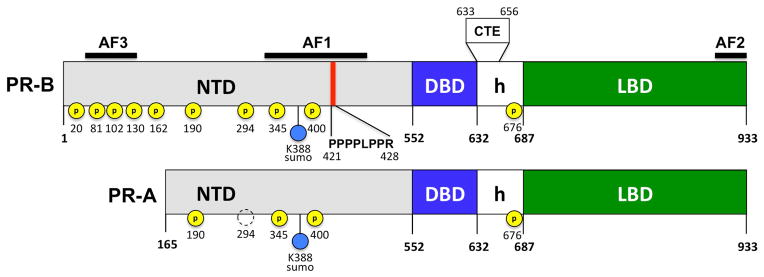
PR-B is the full-length receptor isoform; PR-A lacks 164 amino acids at the N-terminus. Ligand binding domain (LBD), DNA binding domain (DBD), amino terminal domain (NTD), hinge (h), carboxyl terminal extension (CTE), transcriptional activation functions AF1, AF2, AF3. Polyproline motif (as 421–427) that binds the SH3 domain of the tyrosine kinase c-Src, serine phosphorylations (P), sumolylation (SUMO).
Because they are conserved domains, high resolution structures have been determined for isolated LBDs and DBDs of numerous steroid/nuclear receptors. This information has provided important insights into mechanisms of sequence specific DNA binding, ligand binding and how steroid hormone agonists and antagonists regulate interaction of co-activators or co-repressors with AF2. This paper will focus on studies of two naturally unstructured regions of PR, the NTD and hinge region located between the DBD and LBD. These non-conserved domains are less well studied and understood than other domains, yet they are important multi-functional regions essential for the activity of the receptor in their full-length context. We will review work from our laboratories and others on the NTD and hinge of PR as well as related studies with other steroid/nuclear receptors.
2. Ligand binding domain (LBD)
The ligand binding domain (LBD) of steroid/nuclear receptors consists of a common fold comprised of up to 12 helices and a short β-turn arranged in a three layer anti-parallel helical sandwich. The numerical convention of helices 1–12 is based on the first LBD structure RXRα (Bourguet et al., 2000). The GR subfamily of steroid receptors that includes GR, PR, AR and MR, lacks helix 2. Some additional structural features of PR that differ from other receptor LBDs are: helices 10 and 11 are contiguous and therefore are classified as a single helix, the dimer interface is less extensive than other LBDs, and the C-terminal helix 12 has additional residues that pack against helix 10/11 as a loop structure (Madauss et al., 2007; Raaijmakers et al., 2009; Williams and Sigler, 1998). As a general mechanism, helix 12 is dynamic and stabilizes in a position with the other helices to form a hydrophobic cleft that binds LXXLL motifs of SRC family co-activators when occupied by hormone agonist (Bourguet et al., 2000; Nettles et al., 2007). Antagonists bind to the LBD in a conformation that displaces helix 12 and disrupts or prevents formation of the AF2 interaction surface (Brzozowski et al., 1997; Shiau et al., 1998). Early biochemical experiments supported this model for PR. PR bound to agonist adopted a distinct conformation in the C-terminal tail from that of PR bound to the antagonist RU486. This was shown by distinct limited protease digestion patterns and by a monoclonal antibody to the C-terminal tail of PR that only recognizes PR bound to RU486 and not to progesterone (Allan et al., 1992; Vegeto et al., 1992; Weigel et al., 1992). Also, truncation of the C-terminal tail of PR, as well as point mutations, resulted in loss of progesterone binding while RU486 binding was retained, but now acted as a strong agonist (Vegeto et al., 1992; Xu et al., 1996). More recently crystal structures of the PR LBD bound to the antagonists asoprisnil and RU486 (mifepristone) have been reported (Madauss et al., 2007; Raaijmakers et al., 2009). In the asoprisnil structure, helix 12 was displaced and the LBD was bound to a peptide derived from the co-repressor SMRT, resulting in stabilization of helix 12 in the antagonist conformation (Madauss et al., 2007). The RU486 structure was determined by exchange of RU486 with progestin agonist in an existing PR LBD crystal. The crystal packing forces restricted the position of helix 12, resulting in RU486 binding to the PR LBD in an agonist conformation, demonstrating that displacement of helix 12 is not required for binding RU486 (Raaijmakers et al., 2009). Data analysis also showed that helix 12 was more flexible with RU486 bound. These results indicate that RU486 does not induce a single fixed antagonist conformation of helix 12 but rather alters a dynamic equilibrium of helix 12 resulting in destabilization of the agonist conformation.
3. DNA binding domain (DBD)
The DNA binding domain is bipartite, consisting of a conserved zinc finger core of 66 residues and a variable C-terminal extension (CTE) of ~40 amino acids. A boundary between the core PR DBD and CTE is marked by a Gly631, Met632 sequence that is conserved across all nuclear receptors (Figures 3 and and5).5). The core DBD contains two α-helices and two zinc-binding modules that stabilize the globular structure of the domain (Khorasanizadeh and Rastinejad, 2001; Zilliacus et al., 1995). Three residues at the beginning of the DNA recognition helix 1 (P box) play an important role in sequence recognition and discrimination between HREs. The C-terminal zinc-binding module contains a D box which is important for mediating DNA binding-dependent dimerization of the DBD. The consensus recognition hormone response element (HRE) for the glucocorticoid receptor (GR) subgroup of steroid receptors (GR, AR, MR and PR) contains an inverted repeat hexanucleotide sequence 5′-AGAACA-3′ separated by a 3 nucleotide spacer. ER binds to a slightly different consensus hexanucleotide HRE sequence, 5′-AGGTCA-3′ (Umesono and Evans, 1989).
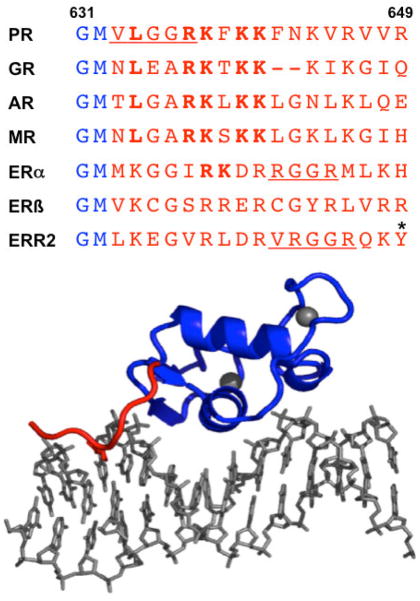
All steroid receptors except ERβ have a conserved RK sequence in a similar position in the CTE as the Arg637/Lys638 residues of PR that interact in the minor groove. Crystal structure of PR DBD showing interaction of Arg637/Lys638 of CTE with the minor groove. View is a monomer subunit bound to a PRE half-site. (Adapted from Molecular Endocrinology 20:3042, 2006)
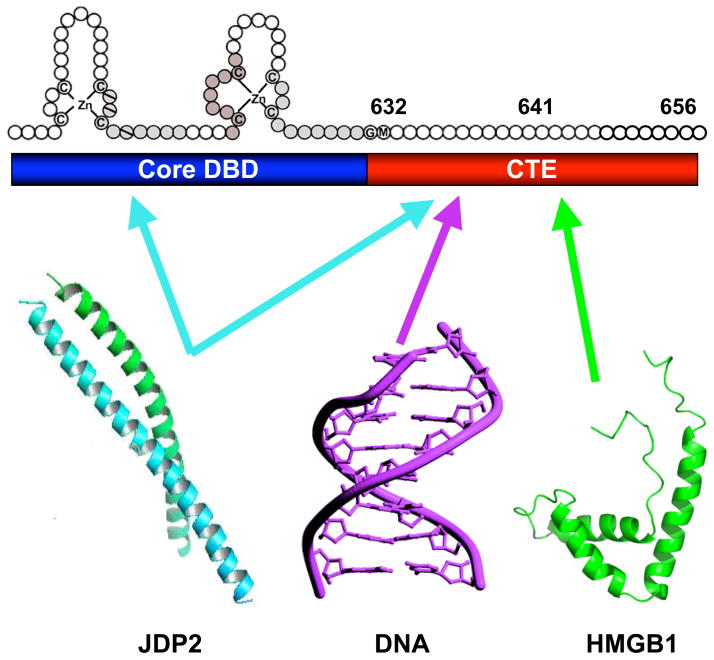
Distinct as well as overlapping residues in the CTE mediate interaction with different partners; aa 632–641 is most important for interaction with DNA and JDP-2, aa 642–656 is more important for interaction with HMGB1. The CTE is capable of adopting distinct conformations dependent on the nature of the interacting partner, and JDP-2-induced structural changes in the CTE are proposed to mediate coupling with the disordered NTD of PR. Adapted from Journal of Biological Chemistry 284: 24422, 2009.
In the PR crystal structure the DBD core was bound to a consensus inverted repeat progesterone response element (PRE) as a head-to-head dimer with helix 1 of each subunit making base specific contacts in the major groove of the PRE hexamers (Roemer et al., 2006) (Figure 2). Base specific contacts are nearly identical to those reported for GR and AR and include hydrogen bonding of Lys588 and Arg593 with Gua6-6-N7 and Gua3-N7 and - O6, and Van der Waals interactions of Val589 with the Cγ methyl of Thy4-C5 in the PRE (Luisi et al., 1991; Shaffer et al., 2004). These contacts are stabilized by a number of direct and water mediated phosphate backbone interactions. Similar to core DBDs of other steroid receptors, helix 2 is perpendicular and above helix 1 and contributes to the overall folding of the domain. The PR DBD contains a short unique helix 2′ that extends from the C-terminus of helix 2. Helix 2′ contributes to the dimer interface and mediates protein-DNA interactions with the phosphate backbone (Roemer et al., 2006). The dimer interface consists of direct and water mediated contacts between residues in the D box (aa 604 to 608) and AR structures, and is further enhanced by interactions outside of the D box including a unique water-mediated contact between Lys617 in helix 2′ of the opposing dimer subunits (Roemer et al., 2006). DNA bound to PR resembled B-form DNA except for the width of the minor groove across the 3N spacer (3.6Å) that was well below the mean (9.6Å) for B-form DNA (Roemer et al., 2006). Other steroid receptor DBD-DNA structures have also exhibited a compressed minor groove width but this is most distorted in the PR-DNA complex. Whether differences in minor groove width reflect intrinsic abilities of different steroid receptors to distort DNA structure is not known. Receptor-induced distortion and directional bending of DNA has been detected for different steroid receptors in solution by circular permutation gel shift assays (Petz et al., 1997; Prendergast et al., 1996; Schultz et al., 2002). An effect on DNA conformation appears to be a general property of steroid receptor-DNA binding that may have a functional role in facilitating the assembly of multi-protein complexes at enhancers or promoters of target genes.
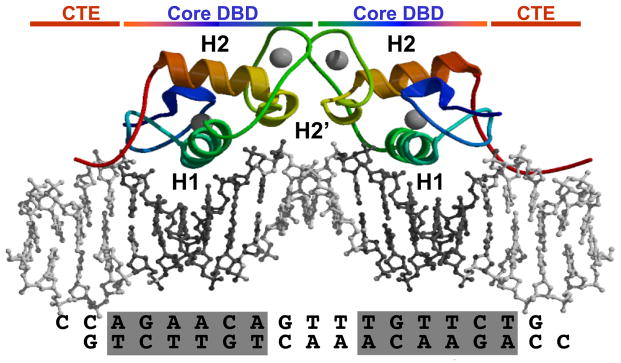
Core DBDs in the dimer are in rainbow coloring from blue (N-terminus) to red (C-terminus). The CTE is red and zinc molecules and DNA are grey. Helix 1 (H1) is green, Helix 2 is red and helix 2′ (light green) extends off the C-terminus of H1. The sequence of the PRE double stranded DNA is indicated at the bottom. Image was made in Pymol. (Adapted from Molecular Endocrinology 20: 3042, 2006)
4. C-terminal extension (CTE) of the DNA binding domain
4.1. Binding to the minor groove flanking HREs of target DNA
The region between the DBD and the LBD, traditionally known as the hinge (Figure 1), is poorly conserved at the primary amino acid sequence level but shares common features among steroid receptors including an enrichment of basic residues and a nuclear localization signal (NLS) for translocation of receptors from the cytoplasm to the nucleus. Work from our lab and others has shown the hinge is much more than a flexible linker containing an NLS. This is a multifunctional region that participates in DNA binding, mediates interaction with co-regulatory proteins and dimerization of receptors, and is a site for post-translational modifications including phosphorylation, acetylation and methylation (Claessens et al., 2008; Daniel et al., 2010; Faus and Haendler, 2006; Fu et al., 2003; Fu et al., 2000; Haelens et al., 2007; Haelens et al., 2003; Hill et al., 2009; Kim et al., 2006; Le Romancer et al., 2010; Melvin et al., 2004; Melvin et al., 2002; Roemer et al., 2008; Roemer et al., 2006; Tanner et al., 2010; Ward and Weigel, 2009). The hinge has been more aptly termed the carboxyl terminal extension (CTE) of the DBD because it directly participates in receptor DNA binding. Structure studies with class II nuclear receptors for non-steroidal ligands, and orphan nuclear receptors, were the first to observe that the CTE interacts with the minor groove outside the HRE to extend the protein-DNA interface beyond the core DBD interactions (Khorasanizadeh and Rastinejad, 2001). Mutational analysis further demonstrated that CTE–DNA interactions enhance stability and DNA binding affinity. The CTE of receptors for thyroid hormone (TR), retinoids (RAR, RXR) and vitamin D (VDR) form a single α-helix, termed a T or A box, which can bind to DNA in two different modes. In crystal structures of TR and VDR, the CTE forms a single α-helix that projected across the DNA minor groove between HRE half-sites and makes extensive non-specific contacts with the DNA backbone (Hsieh et al., 1995; Hsieh et al., 1999; Quack et al., 1998; Rastinejad et al., 1995; Shaffer and Gewirth, 2002). In NMR structures of retinoid X receptor (RXRα) in the absence of DNA the CTE is an α-helix, but when complexed to DNA forms an extended structure indicating the CTE under these conditions unfolds when it interacts with DNA(Lee et al., 1993; Zhao et al., 2000). The CTE of the orphan receptors RevErb, NGF1-B and ERR-2 lie along the floor of the minor groove in an extended loop conformation and contain a short conserved sequence termed the GRIP box (VRXGRIPK, where X is a Phe, Arg or Gly), which makes minor groove contacts with 5′ flanking sequences of each HRE half-site (Zhao et al., 1998). Interaction of the GRIP box appears to provide additional specificity for orphan receptors that bind to extended HRE half-sites enabling them to distinguish 5′ flanking trinucleotide sequences (Gewirth and Sigler, 1995; Zhao et al., 1998). As further indication that the CTE can undergo a conformational change when bound to DNA, the CTE of ERR-2 in the absence of DNA is unstructured (Gearhart et al., 2003; Sem et al., 1997) but makes stabilizing intra-molecular interactions with the core DBD when bound to the minor groove. The structure of the full length PPAR-γ-RXR-α heterodimer complexed with DNA revealed previously unknown interactions between the LBD and DBD and also emphasized the importance of the CTE (Chandra et al., 2008). The CTE of the PPAR-γ subunit of the heterodimer interacts with DNA and is a major determinant of 5′-3′ polarity of the heterodimer while the CTE of the RXR-α subunit mediates protein contact with the PPAR-γ DBD.
The CTE present in the crystal structure of the PR DBD formed an extended loop that interacted with the minor groove flanking either side of the PRE half-sites (Figure 2). The PR CTE resembled that of class II and orphan nuclear receptors by extending the protein DNA interface formed by the core DBD. Arg637 of the CTE was inserted into the minor groove and made a base specific contact through hydrogen bonding to a cytosine flanking the PRE. The next residue, Lys638, hydrogen bonded with Arg637 resulting in the side chain of Lys638 making phosphate contacts that stabilize the CTE-DNA interaction (Roemer et al., 2006). Mutational analysis confirmed the functional importance of these minor groove interactions. Double alanine substitutions for Arg637/Lys638 reduced the binding affinity of PR DBD dimers for palindromic PRE DNA by several fold, but eliminated binding of PR DBD monomers to PRE half-sites (Roemer et al., 2006). This requirement for binding to PRE half-sites suggests that the CTE plays an important role in receptor recognition of natural target genes that frequently contain multiple weak PRE half-sites. We propose that the CTE interface with the minor groove flanking the PRE may be essential for recognition of the full repertoire of target genes regulated by PR in vivo.
Although the CTE is not conserved, Arg637/Lys638 of PR that interacts with the minor groove is conserved in position of all steroid receptor CTEs, except for ERβ (Figure 3). Recent crystallography structures of the GR DBD/DNA complex also observed an interaction of the GR CTE with the minor groove flanking the GRE. Interaction is mediated by Arg510 located in the same position as Arg637 in the CTE of PR, and occurs in a similar manner with the minor groove except that the CTE formed a short α-helix (termed helix 3) rather than an extended loop to orient the position of Arg510 (Meijsing et al., 2009). Similar to results with PR, R510A mutation reduced the GR-DBD binding affinity for GRE DNA (Meijsing et al., 2009). Additional lysine residues in the GR CTE structure projected across the minor groove made additional phosphate backbone contacts that were not present in the PR CTE structure. The GR subgroup of receptors that recognize the same hexanucleotide HRE, contain a conserved RK sequence within an RKXKK basic motif. Deletion of the motif and double mutations of RK in AR also reduced DNA binding in vitro, and this motif was shown to be essential for binding to direct repeat AREs that are present on endogenous AR target genes (Tanner et al., 2010). Domain swapping of the CTE between ERα and ERβ, and deletion mutation experiments have shown that the CTE is important for the higher binding affinity of ERα than ERβ for consensus inverted repeat EREs, and is essential for both ER subtypes to bind to weak ERE half-sites (Melvin et al., 2004). These data taken together suggest that steroid hormone receptors contain a common RK sequence within a basic motif in the CTE that interacts with the minor groove flanking the HRE.
4.2. CTE is a binding site for co-regulatory proteins
Although the most widely studied transcriptional co-regulatory proteins are those that bind with AF2 in the LBD, several co-regulatory proteins have been identified that interact with the DBD of steroid receptors in a manner dependent on the CTE. These include the small nuclear RING finger protein (SNURF), the UBc9 and PIAS proteins responsible for sumolyation of AR, the BAF57-containing SWI/SNF chromatin remodeling complex, and the histone acetyltransferase pCAF (Blanco et al., 1998; Link et al., 2005; Poukka et al., 2000a; Poukka et al., 2000b). Novel ERα co-regulatory proteins have also been identified through biochemical isolation of ERα-ERE complexes including proteins involved in DNA replication, protein folding, translation initiation and elongation, apoptosis, oxidative stress response and base excision DNA repair (Schultz-Norton et al., 2008). Co-repressors have also been identified that interact with the CTE of AR including SMRT, ARR19 and Pod-1 that act by recruitment of histone deacetylases to target genes in vivo (Hong et al., 2005; Jeong et al., 2004; Liao et al., 2003).
Post-translational modifications in the CTE have been identified to be involved in modulating protein interactions with steroid receptors. Methylation of Arg260 in the CTE of ERα, by protein arginine methyltransferase PRMT1, was shown to be required for physical interaction of ER with the p85 subunit of PI3K and with the c-Src tyrosine kinase and for mediating estrogen-induced activation of Src/FAK and p85/Akt signaling pathways involved in proliferation and cell survival (Le Romancer et al., 2010). Acetylation of lysine residues in the CTE of AR has been reported to facilitate or impair AR recruitment of transcriptional activators or repressors (Fu et al., 2003; Fu et al., 2000). Acetylation of Lys266 and Lys268 in the CTE of ERα by p300 was shown to enhance sequence specific DNA binding activity in vitro and estrogen-dependent transcriptional activity of ERα (Kim et al., 2006). Whether acetylation acts by modulating ER interaction with other co-regulatory proteins was not reported (Kim et al., 2006). Two co-regulatory proteins have been identified that enhance PR transcriptional activity through interaction with the CTE including chromatin associated high mobility proteins-1 and 2 (HMGB1/2) and Jun dimerization protein 2 (JDP-2).
4.2.1. High mobility group B proteins (HMGB)
The HMG proteins are a family of functionally related non-histone chromosome associated proteins that contain a structurally conserved HMG box that binds to DNA structures without sequence specificity and can induce distortions in DNA from B-form conformation (Bianchi and Agresti, 2005; Hock et al., 2007). Interaction partners and protein interaction motifs for HMGB proteins have also been defined (Dintilhac and Bernues, 2002). HMGB1 and closely related HMGB2 have been implicated to have a regulatory role in several nuclear processes including V(D)J recombination, DNA replication, transcriptional enhancesome assembly, nucleosome remodeling, transcription elongation and enhancement of sequence specific transcription factor binding to target DNA response elements (see reviews) (Bianchi and Agresti, 2005; Hock et al., 2007). Enhancement of sequence specific DNA binding has been described for Hox proteins, octamer factors, p53 and p73, rel family members and all classes of steroid hormone receptors (Bianchi and Agresti, 2005; Boonyaratanakornkit et al., 1998; Verrijdt et al., 2002). HMGB1 knockout mice die shortly after birth primarily due to glucose metabolism deficiencies; thus, whether HMGB1 has a role for sex steroids in reproduction in vivo cannot be tested by this approach. However, thymocytes from HMGB1 null mice have an impaired glucocorticoid induced apoptosis response indicating a role of HMGB1 for GR function in vivo (Calogero et al., 1999). As further evidence of a role for HMGB1 in steroid hormone receptor action in vivo, interaction of HMGB1 and GR in response to glucocorticoids has been shown to occur on chromatin and on the promoter of an MMTV target gene array in live cells as detected by FRET analysis (Agresti et al., 2005).
The CTE of PR is a protein interaction site for HMGB1 and this interaction has been shown to be essential for HMGB-induced enhancement of PR binding affinity for PREs in vitro and for stimulation of transcriptional activity of PR in intact cells (Melvin et al., 2002; Roemer et al., 2008). A CTE peptide in a sequence specific manner inhibited binding of HMGB1 to PR indicating the CTE alone is sufficient for binding. A minimal region of the CTE required for binding was mapped by mutational analysis to aa 641–651 (Roemer et al., 2008). HMGB1 does not interact with the CTE of class II nuclear receptors for non-steroidal hormones such as TR, RXR, and RAR and does not enhance their DNA binding affinity or transcriptional activity (Boonyaratanakornkit et al., 1998; Melvin et al., 2002; Verrijdt et al., 2002). As further evidence of the essential nature of the CTE for physical and functional interaction of PR with HMGB1, domain swapping of the CTE between PR and class II nuclear receptor resulted in a switch of functional response to HMGB1 (Melvin et al., 2002). HMGB1 did not interact with or stimulate DNA binding activity of a chimeric PR containing the CTE of TR, whereas a chimeric TR containing the CTE of PR exhibited a gain of functional response to HMGB1. Domain swapping and deletion mutation experiments also revealed that class II nuclear receptor DBDs have a higher intrinsic DNA binding affinity than steroid receptors for their respective DNA response elements that is attributable to the CTE. The lower DNA binding affinity of steroid receptor DBDs can be increased to that of class II receptors by addition of HMGB1, indicating that high affinity binding of steroid receptor DBDs requires an auxiliary protein such as HMGB1 (Melvin et al., 2002).
The mechanism by which HMGB1 increases the DNA binding affinity of PR for specific DNA has been explored but remains incompletely defined. HMGB1 is not a stable component of the enhanced high affinity PR/DNA complex in vitro indicating that its interaction is too transient to be detected by biochemical methods. This is a common observation with other steroid receptors and sequence specific transcription factors that exhibit enhanced DNA binding in the presence of HMGB1 (Das et al., 2004; Melvin et al., 2004; Melvin et al., 2002). Point mutations in hydrophobic residues in HMGB1 that reduced binding to non-specific DNA and eliminated DNA bending activity, had no effect on protein interaction of HMGB1 with the PR CTE, or on the ability of HMGB1 to stimulate DNA binding activity of PR in vitro (Roemer et al., 2008). Thus, HMGB1 does not appear to affect PR-DNA binding affinity by manipulating structure of the target DNA, but requires protein interaction with the CTE. Binding of HMGB1 to the PR CTE was inhibited by addition of PRE probes but not by unrelated DNA in vitro indicating that interaction of the CTE with DNA and HMGB1 is mutually exclusive (Roemer et al., 2008).
In addition to these interactions, the CTE appears to mediate a repressive effect on the DNA binding activity of the core DBD. Truncation of the CTE in the context of a PR DBD construct resulted in an increased DNA binding affinity while addition of a peptide corresponding to sequences in the CTE also enhanced DNA binding affinity in a manner dependent on the presence of the CTE in the PR DBD construct (Melvin et al., 2002; Roemer et al., 2008). The CTE peptide had no effect on DNA binding activity of the core DBD alone. These results are consistent with the CTE exerting a repressive effect through an intra-molecular interaction with the core DBD that can be relieved by its removal, or by an external CTE peptide that competes for the interaction (Roemer et al., 2008). In the crystal structure of the orphan receptor ERR-2, the CTE that extends beyond (more C-terminal) the interface with the minor groove formed a loop that interacted with a hydrophobic patch on the core DBD (Zhao et al., 1998). A similar intra-molecular interaction between the CTE and core DBD could occur with PR and account for the increased DNA binding activity caused by deletion of the CTE, or by the addition of a CTE peptide. PR structures with additional CTE sequence will be needed to confirm this hypothesis. A repressive effect of the CTE on DNA binding has similarly been observed for the androgen (AR) and estrogen receptors (Haelens et al., 2007; Melvin et al., 2004).
Since HMGB1 is not stably associated with the receptor/DNA complex, the increase in PR-DNA binding affinity is most likely mediated through additional PR-DNA contacts. We propose that the CTE is in dynamic equilibrium between different conformations and can interact with the core DBD, other proteins or the minor groove of DNA. Transient protein interaction with HMGB1 shifts the equilibrium to favor a conformation of the CTE that is optimal for interaction with DNA minor groove flanking the PRE (Figure 4). Although HMGB1 did not alter sites or the region of protection in an ER/ERE complex by DNase I and exonuclease footprinting, these methods are not expected to detect additional interactions in the minor groove (Das et al., 2004).
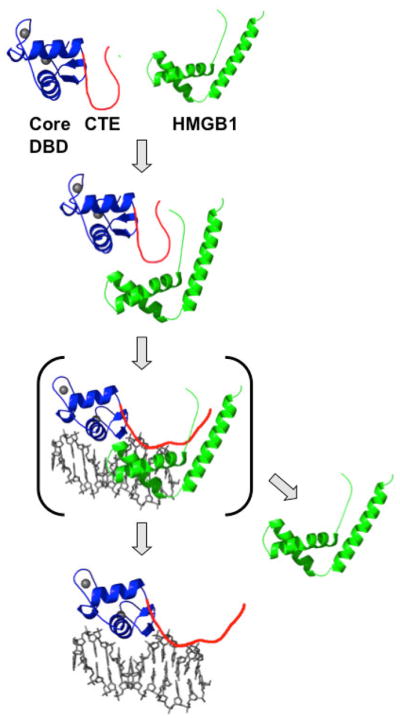
HMGB1 (green) interacts with the CTE (red) of PR (core DBD in blue) and disrupts a repressive intra-molecular interaction between the CTE and core DBD. This promotes CTE binding to the DNA minor groove flanking the PRE. HMGB dissociates and may affect RNA pol II and general transcription factors in down stream steps in the process of steroid receptor-mediated gene transcription. Adapted from Nucleic Acids Research 36:3663, 2008.
What is the physiological role for this interaction of HMGB1 with steroid receptors? Studies with ER demonstrated that HMGB1 had a greater fold enhancement of receptor binding to weak consensus ERE half-sites and divergent EREs than to consensus inverted repeat EREs that bind with the highest affinity (Das et al., 2004). HMGB1 raised the binding affinity for weak EREs to that of consensus EREs. Many, if not most, target genes identified by genomic screening approaches contain HRE half-sites. This occurrence indicates that HMGB1 and the CTE broaden the DNA binding specificity of steroid receptors and may have an essential role in receptor recognition of endogenous steroid receptor target genes. As further evidence that HMGB1 and CTE can affect target gene specificity, a randomized oligonucleotide binding selection assay with the PR DBD-CTE as the target, resulted in an enrichment of AT sequences flanking the 3′ PRE in the presence of HMGB1 as compared with PR alone (Roemer et al., 2008). This is reminiscent of orphan nuclear receptor binding to extended HRE half-sites where the CTE has a preference for AT rich flanking trinucleotide sequences (Zhao et al., 1998).
HMGB1 has other functions separate from its affect on facilitating receptor-DNA binding and recognition of non-consensus HREs. Although mutations in the DNA intercalating residues of HMGB1 were not required to enhance PR DNA binding in vitro, they did reduce HMGB1 enhancement of the transcriptional activity of PR in cells (Roemer et al., 2008). These same mutations also reduced interaction between GR and HMGB in chromatin of live cells as detected by FRET (Agresti et al., 2005). HMGB has been reported to interact with TBP at the initiation complex and to be a component of elongation complexes associated with active RNA pol II on transcribed genes (Das and Scovell, 2001; Guermah et al., 2006). Thus, the DNA intercalating and DNA bending activities of HMGB may be needed for steps of gene transcription down stream of DNA binding including initiating and elongation.
4.2.2. Jun dimerization protein 2 (JDP-2)
JDP-2 was originally identified as a repressor of AP-1 that functions by competing for heterodimerization of c-Jun with c-Fos, or by recruiting histone deacetylases (Aronheim et al., 1997; Jin et al., 2002). JDP-2 has a leucine zipper and a basic amino acid binding domain common to AP-1 transcription factors, but lacks an N-terminal activation domain. It was later identified in a yeast two hybrid screen as a PR interacting protein that enhances transcriptional activity of PR (Edwards et al., 2002). JDP-2 has also been reported to have histone chaperone activity, to be involved in nucleosome assembly, and to inhibit core histone acetylation by the co-activator p300 (Jin et al., 2006). Other bZIP proteins have been reported to function as steroid/nuclear receptor co-activators including XBP-1, which co-activates ERα (Ding et al., 2003) and GT198, a tissue-specific co-activator identified to stimulate activity of several nuclear receptors including ERα and β, TRβ1, AR, GR, and PR (Ko et al., 2002). Interaction of GT198 was reported to be mediated by the DBD of steroid receptors.
Protein binding between PR and JDP-2 is directly mediated by the DBD of PR and the basic domain of JDP-2, and occurs independent of progesterone. This has been detected by pull-down and co-IP assays from cells. JDP-2 does not affect PR-DNA binding and is not detected as a ternary complex with PR and PRE DNA in vitro. However, by chromatin immunoprecipitation (ChIP) assay, JDP-2 is recruited with PR to target gene promoters in a manner stimulated by progesterone (Wardell et al., 2002). JDP-2 enhancement of PR function occurs primarily by stimulating the transcriptional activity mediated by the NTD, with equal effects on the NTD of PR-A and PR-B (Wardell et al., 2002; Wardell et al., 2005). Data from circular dichroism (CD) spectroscopy demonstrated that JDP-2 binding to a two domain PR NTD-DBD construct increased helical content in the NTD by 2 fold (~ 30 to 60%) and the thermostability of PR from a TM of 42°C to 58°C. Partial proteolysis confirmed that JDP-2 binding to PR altered structural conformation in the NTD (Wardell et al., 2005). The effect of JDP-2 on PR NTD structure correlated with transcriptional activity. Other proteins including HMGB1 and closely related JDP-1 that bind to PR DBD did not affect structure or activity of the PR NTD (Wardell et al., 2005). Functional mutagenesis experiments failed to define a single minimal region of the NTD required for JDP-2 stimulation of PR transcriptional activity. Instead, a broad region between aa 327 to 450 was required that appeared to consist of multiple elements. JDP-2 did not stimulate transcriptional activity of chimeric PR constructs consisting of the PR DBD linked to NTDs of other nuclear receptors, or to the GAL4 activation domain. These results indicate that a specific inter-domain communication between PR DBD and its own NTD is required for the effect of JDP-2 and that the DBD plays an active role in modulating activity of the NTD (Wardell et al., 2005). The AF1 of PR was initially defined as aa 456–546 of the NTD based on autonomous activity when regions of the NTD were linked to a heterologous GAL4 DBD (Meyer et al., 1992). However, this region is dispensable for JDP-2 effects on receptor activity when deleted in the context of full length PR (Wardell et al., 2005). These data taken together indicate that AF1 represents multiple elements in the NTD that require protein induced folding. How more ordered structure in the PR NTD induced by JDP-2 alters transcriptional activity is not known. Because it lacks intrinsic transcriptional activity, JDP-2 induced folding of the NTD is presumed to produce protein interaction surfaces for other co-activators. Further experiments will be needed to determine the effects of JDP-2 on interaction of other protein or protein complexes with the NTD.
The progesterone antagonist RU486 can exhibit partial agonist activity under certain conditions, and this is known to require the NTD and to be mediated primarily by PR-B (Meyer et al., 1992). Experiments with JDP-2 provide insights into the mechanism by which NTD can mediate partial agonist activity of RU486. Ectopic expression of JDP-2 potentiated the partial agonist activity of the antagonist RU486, enabling RU486 to induce substantial activation of PR that does not occur under most conditions (Wardell et al., 2010). Mutagenesis experiments revealed that a single region of the NTD (aa 323–427) was required for JDP-2 potentiation of RU486 activity and that phosphorylation of Ser400 within this region was also needed for maximal response. An S400A mutation of PR abrogated JDP-2 potentiation of RU486 agonist activity, but had no effect on agonist-dependent activity of PR in the presence or absence of JDP-2. An S400D PR mutation that retains negative charge character had minimal effect (Wardell et al., 2010). These results suggest that the partial agonist activity of RU486 is mediated by formation of a conformation, and presumably a unique interaction surface in the NTD, that is distinct from that of receptor bound to hormone agonist. These data also support the concept that ligand binding can affect the conformation and activity of the NTD. Since partial agonist activity of RU486 mapped to a single short element in the NTD vs. hormone agonist activity that appears to require multiple elements over a broader region, raises the possibility that folding between multiple elements of the NTD accounts for the higher activity of hormone agonists as compared to the weaker partial agonist activity of RU486.
NMR spectroscopy of C13-N15 labeled PR DBD-CTE and perturbation of chemical shifts was used as an approach to define the amino acids in PR that bind JDP-2 (Hill et al., 2009). The greatest change in chemical shifts occurred in the core DBD second zinc finger and in the CTE between aa 635–638. Point mutations revealed that Arg637/Lys638 in the CTE are the most critical amino acids for JDP-2 binding in vitro and for JDP-2 enhancement of progesterone-dependent PR transcriptional activity in cell co-transfection assays. Peptide competition and mutation analysis confirmed the requirement of the CTE between aa 632–641 for binding JDP-2 and domain swapping confirmed the functional requirement of the CTE for transcriptional co-activation of PR by JDP-2 in cells (Hill et al., 2009). Although point mutations in the Arg637/Lys638 abrogated JDP-2 binding and functional co-activation of PR, progesterone-dependent transcriptional activity, in the absence of JDP-2, was increased indicating these residues have an overall repressive effect on activity of the receptor (Hill et al., 2009). Arg637/Lys638 are the same residues in the crystal structure that insert into the minor groove of DNA and are required for efficient binding of PR to weak PRE half-sites. A similar repressive function of the CTE on hormone-dependent transcriptional activity has been observed for AR, GR and was reported independently for PR. Substitutions for positively charged residues in the RKXKK motif in the CTE of AR resulted in increased transcriptional activity that correlated with an increased intra-nuclear mobility of the receptor and presumably a faster cycling of receptors on and off of target genes enhancers (Tanner et al., 2010). Similar to results with PR, an R510A mutation in the GR CTE increased transcriptional activity of GR towards reporters driven by selective endogenous target gene promoters (Meijsing et al., 2009). Lysine residues in the CTE of PR (K638, K640, K641) were identified to be acetylated in response to progesterone (Daniel et al., 2010). Furthermore, extensive mutational analysis showed that PR acetylation modulates the kinetics, but not the magnitude, of hormone-dependent phosphorylation of receptors, nuclear translocation and activation of early response PR target genes such as c-myc. Interestingly, acetylation of PR also dampened the magnitude of progestin activation of selective slow response target genes such as SGK (Daniel et al., 2010).
These data collectively show that the CTE of PR binds DNA and co-regulatory proteins through overlapping as well as distinct sequences (Figure 5) and mediates a repressive effect on PR DNA binding and transcriptional activity that can be relieved through different mechanisms. To explain the multifunctional properties of this short region of receptors, the CTE is proposed to be dynamic and capable of interacting with multiple macromolecular partners in a transient fashion. Indeed, the predicted NMR structure of the PR DBD-CTE in solution is similar to the crystal structure of the same construct bound to DNA except for the an altered position of the CTE, indicating the CTE adopts a different conformation in solution and bound to DNA (Hill et al., 2009). The CTE has properties ascribed to naturally unstructured polypeptides or intrinsically disordered proteins (IDP) (see below). In multi-domain proteins, IDPs have been proposed to facilitate allosteric coupling between domains propagated through energetic mechanisms as opposed to mechanical pathways (Hilser and Thompson, 2007). The CTE may play such a role in mediating the effects of JDP-2 binding on structural changes in the PR NTD.
5. Thermodynamic properties of PR dimerization and DNA binding
Knowledge of the fundamental properties of PR dimerization and DNA binding are important for understanding the mechanism of PR transcriptional activation. Thermodynamic and hydrodynamic studies of the PR isoforms have provided insights into the gross structural properties of full-length nuclear receptors (Connaghan-Jones et al., 2006; Heneghan et al., 2005). Using analytical ultracentrifugation sedimentation velocity and sedimentation equilibrium analysis, highly purified and functionally homogeneous PR-B was demonstrated to undergo self-association (Heneghan et al., 2005) with an interaction constant (Kd) in the micromolar range (8.8 μM). PR-A has a similar monomer-dimer self association, but occurs at a lower concentration than PR-B with a Kd of 1.08 μM. These data suggest that the additional 164 amino acid residues of the NTD of PR-B have a repressive effect on PR self-association. Interestingly, PR-A as a monomer has a greater frictional ratio than PR-B, indicating a more elongated or larger effective volume, which together with differences observed in limited proteolysis studies of the two receptors supports the idea that the N-terminal residues in PR-B influence other parts of the receptor perhaps allosterically to modulate PR structure and binding activities (Takimoto et al., 2003; Tung et al., 2006). Since PR isoforms have micromolar dimerization constants and levels of receptors in the cell are in the nanomolar suggests that receptors in the cell exist largely in the monomer form.
The energetics of PR-B and PR-A binding to DNA has been dissected using quantitative DNaseI footprint titrations with a single inverted repeat PRE and two tandem PREs, termed PRE2 (Connaghan-Jones et al., 2007; Heneghan et al., 2006). PR-B bound to single PREs with an apparent 12 nM affinity and intra-site cooperativity (Hill coefficient of 1.5). Higher apparent binding affinity (4 nM) and cooperativity (Hill coefficient of 1.8) was observed for inter-site binding between the two tandem PREs and was consistent with previously observed enhancement of binding that occurs with multiple PREs (Heneghan et al., 2006). Together these data were fitted to a thermodynamic binding model that revealed an intrinsic dissociation constant of 81 pM for PR-B dimer binding to a PRE and an intrinsic dissociation constant of 39 nM for PR-B monomers binding to PRE half-sites and an intra-site cooperativity resulting in an apparent Kd of 12 nM. These data lend support to the model for successive monomer binding to a palindromic PRE, as this model was thermodynamically favored over preformed PR dimer binding, perhaps because of additional energy that is required for rearrangement of the PR-B dimer or the DNA binding site (Heneghan et al., 2006). The strong inter-site cooperative interaction between PR-B bound to adjacent PREs was also found to be coupled to energetically unfavorable deformation of PR-B or the promoter DNA. This is in contrast to results for PR-A binding in the same PRE2 system, where PR-A has weaker intrinsic monomer and dimer binding as well as decreased intra-site and inter-site cooperativity. Therefore, amino acid residues unique to PR-B allosterically regulate the energetics of cooperative assembly on promoters (Connaghan-Jones et al., 2007).
PR binding sites on natural target genes are generally composites of near palindromic PREs with adjacent PRE half-sites. The mouse mammary tumor virus (MMTV) promoter, comprised of a single PRE with three tandemly arranged PRE half-sites and a cryptic PRE half-site, was used to examine the binding of PR-A (Connaghan-Jones et al., 2008). In contrast to the weak inter-site cooperativity observed in the PRE2 system, the apparent binding affinity for all MMTV sites has multiple contributions including the modest monomer intrinsic binding affinity, the strong inter-site cooperativity for the tandem half-sites, and additional anti-cooperativity associated with complete saturation of the PRE half-sites. Therefore, PR monomer binding to arrays of tandem PRE half-sites in natural promoters is favorable, cooperative and robust, which leads to synergistic recruitment of co-regulators, such as SRCs that are important for transcriptional activity (Heneghan et al., 2007).
6. Amino-terminal domain (NTD)
Study of intrinsically disordered proteins (IDP) is an emerging field with a growing appreciation of their importance as regulatory domains for cell signaling proteins including transcription factors where IDPs are most frequently located within transcriptional activation domains. IDPs are characterized by a low complexity amino acid composition, often enriched in charged residues, that does not enable spontaneous folding into a globular domain. Thus, IDPs exist as multiple interchanging conformers of extended random coils (Dyson and Wright, 2005; Ferron et al., 2006; Liu et al., 2006). IDPs have a propensity to fold and undergo a disorder-order transition upon binding target proteins by a “coupled folding and binding process,” and they are believed to adopt functionally active conformations by this process. This flexibility is thought to provide an adaptation advantage in that it enables a single regulatory protein to interact with a variety of binding partners. Additionally, the low affinity and high specificity of IDP binding is ideal for reversible transient protein-protein and protein-DNA interactions. Despite the fact that steroid receptor NTDs are not conserved at the amino acid sequence level, it has been proposed they share common structural and mechanistic features as IDPs capable of folding into an active stable conformation upon binding appropriate partner proteins (Kumar and Thompson, 2003; McEwan et al., 2007).
Based on biophysical and hydrodynamic properties, the PR NTD is not globular but adopts an asymmetric elongated rod like shape (Connaghan-Jones et al., 2006; Heneghan et al., 2005; Heneghan et al., 2006). The NTD of PR as well as other SRs is highly susceptible to partial proteolysis, whereas the compact globular LBDs and DBDs are resistant to proteolytic attack (Bain et al., 2000, 2001). Far UV circular dichroism (CD) and fluorescence emission spectroscopy studies have directly shown that NTDs of steroid receptors are largely random coil in solution but undergo cooperative folding and a gain in α helical content in the presence of natural organic osmolytes such as TMAO or in response to binding certain co-regulatory proteins (see reviews) (Kumar and Thompson, 2003; McEwan et al., 2007). Analysis of PR and other steroid receptors using PONDR and GlobPlot algorithms predicts that the NTD contains long stretches of intrinsically disordered protein interspersed with short underlying more structured regions that may represent binding sites for other proteins (McEwan et al., 2007; Tung et al., 2006).
There is a strong preference for post-translation modifications (PTMs) to occur in IDPs (Iakoucheva et al., 2004), and consistent with this, post-translation modifications of steroid receptors including phosphorylation, sumoylation, acetylation, ubiquitination and methylation occurs almost exclusively in the NTD and the naturally unstructured hinge region (or CTE) (Faus and Haendler, 2006; Ward and Weigel, 2009). The segmental flexibility of phosphorylation sites in proteins is required for kinase recognition and for the subsequent binding of phosphorylated residues to target proteins. Steroid receptors have multiple phosphorylation sites that reside largely within the NTD at Ser-Pro motifs (see reviews) (Faus and Haendler, 2006; Ward and Weigel, 2009). Ser-Pro motifs are recognized by the receptor co-activator Pin1, which is a peptidyl prolyl isomerase, and it has been suggested that in addition to causing a change in charge, phosphorylation may also alter receptor structure (Yi et al., 2005). Human PR contains up to10 phosphorylation sites, five are located in the unique PR-B specific region of the NTD, four are in the NTD common to PR-A and PR-B and one site (Ser676) is in the hinge (Figure 1). Although much remains to be learned about the role of phosphorylation of PR, studies have implicated regulation of several functions including receptor turnover, nuclear translocation, transcriptional activity, target promoter selectivity, cross-talk with growth factor signaling pathways and rapid effects of PR on signal transduction pathways that converge on other nuclear transcription factors to regulate gene expression (Daniel and Lange, 2009) and (see reviews) (Faus and Haendler, 2006; Ward and Weigel, 2009). Phosphorylation sites of PR are also differentially modulated at stages of the cell cycle. Ser162 and Ser294 are phosphorylated predominantly during S phase in a manner that correlates with maximal transcriptional activity of PR and recruitment of co-activators to PR target genes suggesting a cell cycle specific role of these phosphorylation events (Narayanan et al., 2005). The only phosphorylation site outside the NTD is Ser676 in the hinge. This is conserved in all steroid receptors and has been most extensively characterized in AR to be involved in regulating nuclear translocation. The Ser294 phosphorylation site has been the most extensively analyzed site in PR. It is located in the NTD common to both PR isoforms, yet phoshorylation of this site is highly preferential for PR-B suggesting that a distinct conformation of PR-B NTD in this region is required for phosphorylation (Clemm et al., 2000). The PR NTD is also modified by sumoylation at Lys388 mediated by the E3 ligase activity of protein inhibitor of activated STAT (PIAS3). Progesterone stimulates rapid sumoylation of Lys388 and mutational analysis demonstrated that this post-translation modification represses transcriptional activity of PR on selected target genes (Abdel-Hafiz et al., 2009; Abdel-Hafiz et al., 2002; Daniel and Lange, 2009; Daniel et al., 2007; Man et al., 2006).
The majority of well characterized nuclear receptor co-activators interact with AF2 in the LBD. Fewer proteins that interact primarily with the NTD and modulate AF1 activity have been characterized. Some examples of AF1 co-activators include p68 (RNA helicase), a DNA repair protein MMS19, an RNA molecule termed SRA (steroid receptor RNA activator), histone acetyltransferase binding to ORC-1 (HBO1), HIC-5, Art-27, AR specific co-activators, and the general transcription factors TBP, TFIIB and TFIIF (see reviews) (Kumar and Thompson, 2003; Li and O’Malley, 2003; McEwan et al., 2007). The NTD can also bind co-repressors such as CHIP, RTA (repressor of tamoxifen transcription activity) and HDAC4 (He et al., 2004; Leong et al., 2005; Martini and Katzenellenbogen, 2003). Of note is the wide variety of interacting proteins and that the NTD is important for cell/tissue type specific activities of steroid receptors. Taken together these properties are consistent with a greater flexibility and range of potential protein interaction partners of the NTD as compared with the conserved well-structured AF2. NTDs of GR, ER and AR have been shown to undergo folding coupled with binding of TBP, CBP, or the RAP74 subunit of TFIIF, in a manner that results in greater α helical content of the NTD (Kumar et al., 2004a; Kumar et al., 2004b; Lavery and McEwan, 2006; Reid et al., 2002; Warnmark et al., 2001). A small core (aa 187–242) of the NTD of GR has been shown to be sufficient for 60–70% of the activity of AF1 and for binding TBP. As determined by NMR spectroscopy, TBP binding generated chemical shift perturbations in the GR core AF1 residues in a region with a propensity to form α helical segments (Kumar et al., 2004b). As evidence of the biological relevance of TBP interaction, an interaction between GR and TBP in live cells was detected by FRET and TBP enhanced the transcriptional activity of GR in cell transfection assays in a manner dependent on the AF1 core (Copik et al., 2006). In cell-free transcription assays with the AF1 of AR linked to a heterologous DBD, TFIIF binding enhanced activity of AF1 (Choudhry et al., 2006). Recent studies to define the mechanism by which the NTD of GR mediates transcriptional activity revealed that phosphorylation of Ser211 in the core AF1 induced secondary and tertiary structure formation that facilitated interaction of AF1 with co-regulatory proteins including TBP, SRC1 and CBP. This study for the first time implicates a role for site-specific phosphorylation in directly modulating the structure of the NTD to adopt a more ordered functionally active conformation (Garza et al., 2010).
TBP also binds to the NTD of PR and induces stable tertiary and secondary (α-helix) structure in a manner that correlates with increased transcriptional activity (Moure, 2009). As assessed by circular dichroism spectroscopy, the NTD of PR exhibited a decrease in random coil and an increase in helical content in response to binding TBP. It also adopted a more ordered tertiary structure in the presence of TBP as determined by protection at multiple sites against proteolysis and by fluorescence emission spectroscopy. Mapping experiments defined a minimal region of the NTD (aa 350–428) that folds in response to binding TBP and point mutations in a predicted short α-helix in this region (aa 384–410) disrupted TBP coupled folding and binding. Mutations in the NTD region that disrupted TBP coupled folding and binding, also reduced TBP enhancement of PR transcription activity (Moure, 2009). These data suggest that similar to GR and AR, PR contains a short α-helical element embedded within the NTD that binds TBP and nucleates folding of the NTD to adopt a more stable structure.
In addition to the well characterized interaction between the LXXLL motif in the nuclear receptor (NR) box of steroid receptor coactivators (SRCs) with AF2 in the C-terminus of steroid receptors, interactions have also been documented between other surfaces of SRCs with the NTD/AF1 (Ma et al., 1999; Metivier et al., 2001; Onate et al., 1998; Webb et al., 1998). As evidence that these interactions are functionally important, full-length SRC-1 bearing mutations in the NR box that selectively disrupts interaction with AF2 are still potent coactivators of steroid receptors (Ma et al., 1999). A hormone agonist-dependent interaction between the amino and carboxyl termini of PR contributes to functional synergy between AF1 and AF2, and SRC-1 has also been demonstrated to enhance N-C terminal interactions (Abdel-Hafiz et al., 2009; Tetel et al., 1999). Whether the interaction of SRCs with AF1 affects folding of the NTD has not been explored.
The NTD is responsible for differences in transcriptional activity of the two PR isoforms since they differ only by unique additional sequences in the NTD of PR-B. The precise mechanism(s) responsible for the distinct activities of PR-A and PR-B is not known. Data suggests the NTD structure of the two isoforms differs and that the B-specific region exerts allosteric effects on conformation and functional activities of other regions of the receptor (Bain et al., 2000, 2001; Tung et al., 2006). Distinct structural conformations of the NTD may also be responsible for the preferential phosphorylation of Ser294 in PR-A (Clemm et al., 2000; Daniel et al., 2007). PR-B contains a third transcription activation domain AF3 that was mapped to residues aa 54–154 in the PR-B specific region of the NTD, and three specific sites were identified that define the minimal AF3 activity; Trp140 and two LXLL motifs termed L1 and L2 (Tung et al., 2001). AF3 contains autonomous transcriptional activity when linked to a heterologous DBD, but its main function is to synergize with AF1 and AF2 in the context of full length PR-B. Support for this comes from experiments where mutations in Trp140 and L1/L2 that abolished autonomous AF3 activity also inactivated full-length PR-B and did not convert PR-B functionally to PR-A based on target gene regulation (Tung et al., 2006). Additionally, AF3 synergy required cooperative interactions of PR with multiple-PRE promoters, was associated with recruitment of SRC-1/p160 co-activator complexes, and required the sumoylation of the NTD (Takimoto et al., 2003; Tung et al., 2006).
In addition to induced folding through protein binding, the structure of the NTD can be stabilized in response to binding DNA via an inter-domain allosteric mechanism. Studies of two domain NTD-DBD polypeptides of AR, GR and PR have shown that DNA binding protected areas of the NTD against proteolytic attack, and CD and fluorescence emission data show a cooperative folding of the NTD in response to DNA binding (Bain et al., 2000, 2001; Kumar et al., 1999). Studies with full-length ER and GR have also shown by proteolysis and other methods that binding to DNA alters the conformation of receptors and co-activator interaction (Hall et al., 2002; Lefstin and Yamamoto, 1998; Wood et al., 2001). Binding to slightly different HRE sequences was observed to induce distinct conformations in the receptor (Hall et al., 2002; Wood et al., 2001) and to direct preferential utilization of co-activators and activation domains (AF1 vs. AF2) (Meijsing et al., 2009). These data indicate that DNA is a sequence specific allosteric ligand that can modulate the activity of receptors bound to specific target genes. X-ray crystallography studies of the GR DBD, complexed with 13 different GRE sequences, observed that structures were indistinguishable except for distinct changes in the conformation of a loop (termed lever arm) that connects the DNA recognition helix 1 and the D box. Change in lever arm conformation has been proposed to be involved in transmitting signals from the DBD to other domains of receptors in response to binding DNA (Meijsing et al., 2009). We propose the CTE that is also conformationally sensitive to DNA and protein interactions may play a similar role (Hill et al., 2009; Roemer et al., 2008).
7. Conclusion
The carboxyl terminal extension (CTE) of the DNA binding domain and the NTD of PR are dynamic regions of the receptor that can adopt multiple conformations and structures depending on the environment of interacting proteins and DNA. Both regions have regulatory roles for multiple receptor functions that appear to relate to the potential to form multiple active conformations. Despite the lack of understanding of the structure/function of the NTD, it accounts for a major proportion of the total transcriptional activity of steroid receptors, is important for cell and target gene specific activities, and it contributes to the relative agonist/antagonist activities of small molecule ligand selective receptor modulators (SRMs). The design of SRMs for clinical use and what we know mechanistically about how SRMs work is based primarily on how they alter conformation and interactions of co-regulatory proteins with AF2 in the LBD. A better definition of the structure/function properties of the NTD is essential for the development of improved SRMs and for advancing our understanding of the molecular mechanism of action of steroid receptors. A high resolution structure of full length steroid receptors and the NTD has eluded the field for a long time. This will likely require decoration of the NTD with appropriate protein and DNA binding partners that promote folding as a stable structure.
Acknowledgments
Portions of the work on PR performed by the authors were supported by NIH/NCI-CA46938 (DPE and MEAC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Hafiz H, Dudevoir ML, Horwitz KB. Mechanisms underlying the control of progesterone receptor transcriptional activity by SUMOylation. J Biol Chem. 2009;284:9099–9108. [Europe PMC free article] [Abstract] [Google Scholar]
- Abdel-Hafiz H, Takimoto GS, Tung L, Horwitz KB. The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J Biol Chem. 2002;277:33950–33956. [Abstract] [Google Scholar]
- Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME. GR and HMGB1 interact only within chromatin and influence each other’s residence time. Mol Cell. 2005;18:109–121. [Abstract] [Google Scholar]
- Allan GF, Leng X, Tsai SY, Weigel NL, Edwards DP, Tsai MJ, O’Malley BW. Hormone and antihormone induce distinct conformational changes which are central to steroid receptor activation. J Biol Chem. 1992;267:19513–19520. [Abstract] [Google Scholar]
- Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–3102. [Europe PMC free article] [Abstract] [Google Scholar]
- Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of the human progesterone A-receptor. Structural analysis and the influence of the DNA binding domain. J Biol Chem. 2000;275:7313–7320. [Abstract] [Google Scholar]
- Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of human progesterone B-receptors: biophysical and biochemical comparison to A-receptors. J Biol Chem. 2001;276:23825–23831. [Abstract] [Google Scholar]
- Ballare C, Uhrig M, Bechtold T, Sancho E, Di Domenico M, Migliaccio A, Auricchio F, Beato M. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol. 2003;23:1994–2008. [Europe PMC free article] [Abstract] [Google Scholar]
- Beato M, Klug J. Steroid hormone receptors: an update. Hum Reprod Update. 2000;6:225–236. [Abstract] [Google Scholar]
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. [Abstract] [Google Scholar]
- Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. [Europe PMC free article] [Abstract] [Google Scholar]
- Boonyaratanakornkit V, McGowan E, Sherman L, Mancini MA, Cheskis BJ, Edwards DP. The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol Endocrinol. 2007;21:359–375. [Abstract] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. [Europe PMC free article] [Abstract] [Google Scholar]
- Boonyaratanakornkit V, Scott MP, Ribon V, Sherman L, Anderson SM, Maller JL, Miller WT, Edwards DP. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol Cell. 2001;8:269–280. [Abstract] [Google Scholar]
- Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21:381–388. [Abstract] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. [Abstract] [Google Scholar]
- Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. [Abstract] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. [Europe PMC free article] [Abstract] [Google Scholar]
- Choudhry MA, Ball A, McEwan IJ. The role of the general transcription factor IIF in androgen receptor-dependent transcription. Mol Endocrinol. 2006;20:2052–2061. [Abstract] [Google Scholar]
- Claessens F, Denayer S, Van Tilborgh N, Kerkhofs S, Helsen C, Haelens A. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal. 2008;6:e008. [Europe PMC free article] [Abstract] [Google Scholar]
- Clemm DL, Sherman L, Boonyaratanakornkit V, Schrader WT, Weigel NL, Edwards DP. Differential hormone-dependent phosphorylation of progesterone receptor A and B forms revealed by a phosphoserine site-specific monoclonal antibody. Mol Endocrinol. 2000;14:52–65. [Abstract] [Google Scholar]
- Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL. Hydrodynamic analysis of the human progesterone receptor A-isoform reveals that self-association occurs in the micromolar range. Biochemistry. 2006;45:12090–12099. [Abstract] [Google Scholar]
- Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL. Thermodynamic analysis of progesterone receptor-promoter interactions reveals a molecular model for isoform-specific function. Proc Natl Acad Sci U S A. 2007;104:2187–2192. [Europe PMC free article] [Abstract] [Google Scholar]
- Connaghan-Jones KD, Heneghan AF, Miura MT, Bain DL. Thermodynamic dissection of progesterone receptor interactions at the mouse mammary tumor virus promoter: monomer binding and strong cooperativity dominate the assembly reaction. J Mol Biol. 2008;377:1144–1160. [Europe PMC free article] [Abstract] [Google Scholar]
- Copik AJ, Webb MS, Miller AL, Wang Y, Kumar R, Thompson EB. Activation function 1 of glucocorticoid receptor binds TATA-binding protein in vitro and in vivo. Mol Endocrinol. 2006;20:1218–1230. [Abstract] [Google Scholar]
- Daniel AR, Gaviglio AL, Czaplicki LM, Hillard CJ, Housa D, Lange CA. The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention. Mol Endocrinol. 2010;24:2126–2138. [Europe PMC free article] [Abstract] [Google Scholar]
- Daniel AR, Lange CA. Protein kinases mediate ligand-independent derepression of sumoylated progesterone receptors in breast cancer cells. Proc Natl Acad Sci U S A. 2009;106:14287–14292. [Europe PMC free article] [Abstract] [Google Scholar]
- Daniel AR, Qiu M, Faivre EJ, Ostrander JH, Skildum A, Lange CA. Linkage of progestin and epidermal growth factor signaling: phosphorylation of progesterone receptors mediates transcriptional hypersensitivity and increased ligand-independent breast cancer cell growth. Steroids. 2007;72:188–201. [Europe PMC free article] [Abstract] [Google Scholar]
- Das D, Peterson RC, Scovell WM. High mobility group B proteins facilitate strong estrogen receptor binding to classical and half-site estrogen response elements and relax binding selectivity. Mol Endocrinol. 2004;18:2616–2632. [Abstract] [Google Scholar]
- Das D, Scovell WM. The binding interaction of HMG-1 with the TATA-binding protein/TATA complex. J Biol Chem. 2001;276:32597–32605. [Abstract] [Google Scholar]
- Ding L, Yan J, Zhu J, Zhong H, Lu Q, Wang Z, Huang C, Ye Q. Ligand-independent activation of estrogen receptor alpha by XBP-1. Nucleic Acids Res. 2003;31:5266–5274. [Europe PMC free article] [Abstract] [Google Scholar]
- Dintilhac A, Bernues J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J Biol Chem. 2002;277:7021–7028. [Abstract] [Google Scholar]
- Dong X, Challis JR, Lye SJ. Intramolecular interactions between the AF3 domain and the C-terminus of the human progesterone receptor are mediated through two LXXLL motifs. J Mol Endocrinol. 2004;32:843–857. [Abstract] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. [Abstract] [Google Scholar]
- Edwards DP, Wardell SE, Boonyaratanakornkit V. Progesterone receptor interacting coregulatory proteins and cross talk with cell signaling pathways. J Steroid Biochem Mol Biol. 2002;83:173–186. [Abstract] [Google Scholar]
- Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60:520–528. [Abstract] [Google Scholar]
- Fernandez-Valdivia R, Mukherjee A, Mulac-Jericevic B, Conneely OM, DeMayo FJ, Amato P, Lydon JP. Revealing progesterone’s role in uterine and mammary gland biology: insights from the mouse. Semin Reprod Med. 2005;23:22–37. [Abstract] [Google Scholar]
- Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. [Abstract] [Google Scholar]
- Fu M, Rao M, Wang C, Sakamaki T, Wang J, Di Vizio D, Zhang X, Albanese C, Balk S, Chang C, et al. Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol Cell Biol. 2003;23:8563–8575. [Europe PMC free article] [Abstract] [Google Scholar]
- Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275:20853–20860. [Abstract] [Google Scholar]
- Garza AM, Khan SH, Kumar R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol. 2010;30:220–230. [Europe PMC free article] [Abstract] [Google Scholar]
- Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol. 2003;327:819–832. [Abstract] [Google Scholar]
- Gewirth DT, Sigler PB. The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex. Nat Struct Biol. 1995;2:386–394. [Abstract] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. [Abstract] [Google Scholar]
- Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F. The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res. 2007;67:4514–4523. [Abstract] [Google Scholar]
- Haelens A, Verrijdt G, Callewaert L, Christiaens V, Schauwaers K, Peeters B, Rombauts W, Claessens F. DNA recognition by the androgen receptor: evidence for an alternative DNA-dependent dimerization, and an active role of sequences flanking the response element on transactivation. Biochem J. 2003;369:141–151. [Europe PMC free article] [Abstract] [Google Scholar]
- Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. [Abstract] [Google Scholar]
- He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, Wilson EM. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J Biol Chem. 2004;279:30643–30653. [Abstract] [Google Scholar]
- Heneghan AF, Berton N, Miura MT, Bain DL. Self-association energetics of an intact, full-length nuclear receptor: the B-isoform of human progesterone receptor dimerizes in the micromolar range. Biochemistry. 2005;44:9528–9537. [Abstract] [Google Scholar]
- Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Cooperative DNA binding by the B-isoform of human progesterone receptor: thermodynamic analysis reveals strongly favorable and unfavorable contributions to assembly. Biochemistry. 2006;45:3285–3296. [Europe PMC free article] [Abstract] [Google Scholar]
- Heneghan AF, Connaghan-Jones KD, Miura MT, Bain DL. Coactivator assembly at the promoter: efficient recruitment of SRC2 is coupled to cooperative DNA binding by the progesterone receptor. Biochemistry. 2007;46:11023–11032. [Abstract] [Google Scholar]
- Hill KK, Roemer SC, Jones DN, Churchill ME, Edwards DP. A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. J Biol Chem. 2009;284:24415–24424. [Europe PMC free article] [Abstract] [Google Scholar]
- Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104:8311–8315. [Europe PMC free article] [Abstract] [Google Scholar]
- Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. [Europe PMC free article] [Abstract] [Google Scholar]
- Hong CY, Gong EY, Kim K, Suh JH, Ko HM, Lee HJ, Choi HS, Lee K. Modulation of the expression and transactivation of androgen receptor by the basic helix-loop-helix transcription factor Pod-1 through recruitment of histone deacetylase 1. Mol Endocrinol. 2005;19:2245–2257. [Abstract] [Google Scholar]
- Hsieh JC, Jurutka PW, Selznick SH, Reeder MC, Haussler CA, Whitfield GK, Haussler MR. The T-box near the zinc fingers of the human vitamin D receptor is required for heterodimeric DNA binding and transactivation. Biochem Biophys Res Commun. 1995;215:1–7. [Abstract] [Google Scholar]
- Hsieh JC, Whitfield GK, Oza AK, Dang HT, Price JN, Galligan MA, Jurutka PW, Thompson PD, Haussler CA, Haussler MR. Characterization of unique DNA-binding and transcriptional-activation functions in the carboxyl-terminal extension of the zinc finger region in the human vitamin D receptor. Biochemistry. 1999;38:16347–16358. [Abstract] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. [Europe PMC free article] [Abstract] [Google Scholar]
- Jeong BC, Hong CY, Chattopadhyay S, Park JH, Gong EY, Kim HJ, Chun SY, Lee K. Androgen receptor corepressor-19 kDa (ARR19), a leucine-rich protein that represses the transcriptional activity of androgen receptor through recruitment of histone deacetylase. Mol Endocrinol. 2004;18:13–25. [Abstract] [Google Scholar]
- Jin C, Kato K, Chimura T, Yamasaki T, Nakade K, Murata T, Li H, Pan J, Zhao M, Sun K, et al. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat Struct Mol Biol. 2006;13:331–338. [Abstract] [Google Scholar]
- Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, Yokoyama KK. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol Cell Biol. 2002;22:4815–4826. [Europe PMC free article] [Abstract] [Google Scholar]
- Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. [Europe PMC free article] [Abstract] [Google Scholar]
- Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26:384–390. [Abstract] [Google Scholar]
- Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol. 2006;20:1479–1493. [Europe PMC free article] [Abstract] [Google Scholar]
- Ko L, Cardona GR, Henrion-Caude A, Chin WW. Identification and characterization of a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. Mol Cell Biol. 2002;22:357–369. [Europe PMC free article] [Abstract] [Google Scholar]
- Kumar R, Baskakov IV, Srinivasan G, Bolen DW, Lee JC, Thompson EB. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J Biol Chem. 1999;274:24737–24741. [Abstract] [Google Scholar]
- Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004a;43:3008–3013. [Abstract] [Google Scholar]
- Kumar R, Thompson EB. Transactivation functions of the N-terminal domains of nuclear hormone receptors: protein folding and coactivator interactions. Mol Endocrinol. 2003;17:1–10. [Abstract] [Google Scholar]
- Kumar R, Volk DE, Li J, Lee JC, Gorenstein DG, Thompson EB. TATA box binding protein induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc Natl Acad Sci U S A. 2004b;101:16425–16430. [Europe PMC free article] [Abstract] [Google Scholar]
- Lavery DN, McEwan IJ. The human androgen receptor AF1 transactivation domain: interactions with transcription factor IIF and molten-globule-like structural characteristics. Biochem Soc Trans. 2006;34:1054–1057. [Abstract] [Google Scholar]
- Le Romancer M, Treilleux I, Bouchekioua-Bouzaghou K, Sentis S, Corbo L. Methylation, a key step for nongenomic estrogen signaling in breast tumors. Steroids. 2010;75:560–564. [Abstract] [Google Scholar]
- Lee MS, Kliewer SA, Provencal J, Wright PE, Evans RM. Structure of the retinoid X receptor alpha DNA binding domain: a helix required for homodimeric DNA binding. Science. 1993;260:1117–1121. [Abstract] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. [Abstract] [Google Scholar]
- Leong H, Sloan JR, Nash PD, Greene GL. Recruitment of histone deacetylase 4 to the N-terminal region of estrogen receptor alpha. Mol Endocrinol. 2005;19:2930–2942. [Abstract] [Google Scholar]
- Li X, O’Malley BW. Unfolding the action of progesterone receptors. J Biol Chem. 2003;278:39261–39264. [Abstract] [Google Scholar]
- Liao G, Chen LY, Zhang A, Godavarthy A, Xia F, Ghosh JC, Li H, Chen JD. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem. 2003;278:5052–5061. [Abstract] [Google Scholar]
- Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, Weissman B, Knudsen KE. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–2215. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. [Europe PMC free article] [Abstract] [Google Scholar]
- Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. [Abstract] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. [Abstract] [Google Scholar]
- Ma H, Hong H, Huang SM, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. [Europe PMC free article] [Abstract] [Google Scholar]
- Madauss KP, Grygielko ET, Deng SJ, Sulpizio AC, Stanley TB, Wu C, Short SA, Thompson SK, Stewart EL, Laping NJ, et al. A structural and in vitro characterization of asoprisnil: a selective progesterone receptor modulator. Mol Endocrinol. 2007;21:1066–1081. [Abstract] [Google Scholar]
- Man JH, Li HY, Zhang PJ, Zhou T, He K, Pan X, Liang B, Li AL, Zhao J, Gong WL, et al. PIAS3 induction of PRB sumoylation represses PRB transactivation by destabilizing its retention in the nucleus. Nucleic Acids Res. 2006;34:5552–5566. [Europe PMC free article] [Abstract] [Google Scholar]
- Martini PG, Katzenellenbogen BS. Modulation of estrogen receptor activity by selective coregulators. J Steroid Biochem Mol Biol. 2003;85:117–122. [Abstract] [Google Scholar]
- McEwan IJ, Lavery D, Fischer K, Watt K. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal. 2007;5:e001. [Europe PMC free article] [Abstract] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. [Europe PMC free article] [Abstract] [Google Scholar]
- Melvin VS, Harrell C, Adelman JS, Kraus WL, Churchill M, Edwards DP. The role of the C-terminal extension (CTE) of the estrogen receptor alpha and beta DNA binding domain in DNA binding and interaction with HMGB. J Biol Chem. 2004;279:14763–14771. [Abstract] [Google Scholar]
- Melvin VS, Roemer SC, Churchill ME, Edwards DP. The C-terminal extension (CTE) of the nuclear hormone receptor DNA binding domain determines interactions and functional response to the HMGB-1/-2 co-regulatory proteins. J Biol Chem. 2002;277:25115–25124. [Abstract] [Google Scholar]
- Metivier R, Penot G, Flouriot G, Pakdel F. Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 alpha-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol. 2001;15:1953–1970. [Abstract] [Google Scholar]
- Meyer ME, Pornon A, Ji JW, Bocquel MT, Chambon P, Gronemeyer H. Agonistic and antagonistic activities of RU486 on the functions of the human progesterone receptor. EMBO J. 1990;9:3923–3932. [Europe PMC free article] [Abstract] [Google Scholar]
- Meyer ME, Quirin-Stricker C, Lerouge T, Bocquel MT, Gronemeyer H. A limiting factor mediates the differential activation of promoters by the human progesterone receptor isoforms. J Biol Chem. 1992;267:10882–10887. [Abstract] [Google Scholar]
- Mote PA, Balleine RL, McGowan EM, Clarke CL. Colocalization of progesterone receptors A and B by dual immunofluorescent histochemistry in human endometrium during the menstrual cycle. J Clin Endocrinol Metab. 1999;84:2963–2971. [Abstract] [Google Scholar]
- Mote PA, Bartow S, Tran N, Clarke CL. Loss of co-ordinate expression of progesterone receptors A and B is an early event in breast carcinogenesis. Breast Cancer Res Treat. 2002;72:163–172. [Abstract] [Google Scholar]
- Moure MC, Callaway C, Kumar R, Edwards DP. Protein induced folding of the intrinsically disordered amino-terminal domain of progesterone receptor controls transcriptional activity of AF1. Annual Endocrine Society Meetings; Washington, D.C. 2009. [Google Scholar]
- Narayanan R, Edwards DP, Weigel NL. Human progesterone receptor displays cell cycle-dependent changes in transcriptional activity. Mol Cell Biol. 2005;25:2885–2898. [Europe PMC free article] [Abstract] [Google Scholar]
- Nettles KW, Bruning JB, Gil G, O’Neill EE, Nowak J, Guo Y, Kim Y, DeSombre ER, Dilis R, Hanson RN, et al. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007;8:563–568. [Europe PMC free article] [Abstract] [Google Scholar]
- Onate SA, Boonyaratanakornkit V, Spencer TE, Tsai SY, Tsai MJ, Edwards DP, O’Malley BW. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. [Abstract] [Google Scholar]
- Petz LN, Nardulli AM, Kim J, Horwitz KB, Freedman LP, Shapiro DJ. DNA bending is induced by binding of the glucocorticoid receptor DNA binding domain and progesterone receptors to their response element. J Steroid Biochem Mol Biol. 1997;60:31–41. [Abstract] [Google Scholar]
- Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci U S A. 2000a;97:14145–14150. [Europe PMC free article] [Abstract] [Google Scholar]
- Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo JJ, Janne OA. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J Cell Sci. 2000b;113(Pt 17):2991–3001. [Abstract] [Google Scholar]
- Prendergast P, Pan Z, Edwards DP. Progesterone receptor-induced bending of its target DNA: distinct effects of the A and B receptor forms. Mol Endocrinol. 1996;10:393–407. [Abstract] [Google Scholar]
- Proietti C, Salatino M, Rosemblit C, Carnevale R, Pecci A, Kornblihtt AR, Molinolo AA, Frahm I, Charreau EH, Schillaci R, Elizalde PV. Progestins induce transcriptional activation of signal transducer and activator of transcription 3 (Stat3) via a Jak- and Src-dependent mechanism in breast cancer cells. Mol Cell Biol. 2005;25:4826–4840. [Europe PMC free article] [Abstract] [Google Scholar]
- Quack M, Szafranski K, Rouvinen J, Carlberg C. The role of the T-box for the function of the vitamin D receptor on different types of response elements. Nucleic Acids Res. 1998;26:5372–5378. [Europe PMC free article] [Abstract] [Google Scholar]
- Raaijmakers HC, Versteegh JE, Uitdehaag JC. The X-ray structure of RU486 bound to the progesterone receptor in a destabilized agonistic conformation. J Biol Chem. 2009;284:19572–19579. [Europe PMC free article] [Abstract] [Google Scholar]
- Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. [Abstract] [Google Scholar]
- Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. 2002;277:20079–20086. [Abstract] [Google Scholar]
- Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem. 2002;277:5209–5218. [Abstract] [Google Scholar]
- Roemer SC, Adelman J, Churchill ME, Edwards DP. Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding. Nucleic Acids Res. 2008;36:3655–3666. [Europe PMC free article] [Abstract] [Google Scholar]
- Roemer SC, Donham DC, Sherman L, Pon VH, Edwards DP, Churchill ME. Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements. Mol Endocrinol. 2006;20:3042–3052. [Europe PMC free article] [Abstract] [Google Scholar]
- Saitoh M, Ohmichi M, Takahashi K, Kawagoe J, Ohta T, Doshida M, Takahashi T, Igarashi H, Mori-Abe A, Du B, et al. Medroxyprogesterone acetate induces cell proliferation through up-regulation of cyclin D1 expression via phosphatidylinositol 3-kinase/Akt/nuclear factor-kappaB cascade in human breast cancer cells. Endocrinology. 2005;146:4917–4925. [Abstract] [Google Scholar]
- Schultz-Norton JR, Ziegler YS, Likhite VS, Yates JR, Nardulli AM. Isolation of novel coregulatory protein networks associated with DNA-bound estrogen receptor alpha. BMC Mol Biol. 2008;9:97. [Europe PMC free article] [Abstract] [Google Scholar]
- Schultz JR, Loven MA, Melvin VM, Edwards DP, Nardulli AM. Differential modulation of DNA conformation by estrogen receptors alpha and beta. J Biol Chem. 2002;277:8702–8707. [Abstract] [Google Scholar]
- Sem DS, Casimiro DR, Kliewer SA, Provencal J, Evans RM, Wright PE. NMR spectroscopic studies of the DNA-binding domain of the monomer-binding nuclear orphan receptor, human estrogen related receptor-2. The carboxyl-terminal extension to the zinc-finger region is unstructured in the free form of the protein. J Biol Chem. 1997;272:18038–18043. [Abstract] [Google Scholar]
- Shaffer PL, Gewirth DT. Structural basis of VDR-DNA interactions on direct repeat response elements. EMBO J. 2002;21:2242–2252. [Europe PMC free article] [Abstract] [Google Scholar]
- Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A. 2004;101:4758–4763. [Europe PMC free article] [Abstract] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. [Abstract] [Google Scholar]
- Skildum A, Faivre E, Lange CA. Progesterone receptors induce cell cycle progression via activation of mitogen-activated protein kinases. Mol Endocrinol. 2005;19:327–339. [Abstract] [Google Scholar]
- Takimoto GS, Tung L, Abdel-Hafiz H, Abel MG, Sartorius CA, Richer JK, Jacobsen BM, Bain DL, Horwitz KB. Functional properties of the N-terminal region of progesterone receptors and their mechanistic relationship to structure. J Steroid Biochem Mol Biol. 2003;85:209–219. [Abstract] [Google Scholar]
- Tanner TM, Denayer S, Geverts B, Van Tilborgh N, Kerkhofs S, Helsen C, Spans L, Dubois V, Houtsmuller AB, Claessens F, Haelens A. A 629RKLKK633 motif in the hinge region controls the androgen receptor at multiple levels. Cell Mol Life Sci. 2010;67:1919–1927. [Europe PMC free article] [Abstract] [Google Scholar]
- Tetel MJ, Giangrande PH, Leonhardt SA, McDonnell DP, Edwards DP. Hormone-dependent interaction between the amino- and carboxyl-terminal domains of progesterone receptor in vitro and in vivo. Mol Endocrinol. 1999;13:910–924. [Abstract] [Google Scholar]
- Tung L, Abdel-Hafiz H, Shen T, Harvell DM, Nitao LK, Richer JK, Sartorius CA, Takimoto GS, Horwitz KB. Progesterone receptors (PR)-B and -A regulate transcription by different mechanisms: AF-3 exerts regulatory control over coactivator binding to PR-B. Mol Endocrinol. 2006;20:2656–2670. [Abstract] [Google Scholar]
- Tung L, Mohamed MK, Hoeffler JP, Takimoto GS, Horwitz KB. Antagonist-occupied human progesterone B-receptors activate transcription without binding to progesterone response elements and are dominantly inhibited by A-receptors. Mol Endocrinol. 1993;7:1256–1265. [Abstract] [Google Scholar]
- Tung L, Shen T, Abel MG, Powell RL, Takimoto GS, Sartorius CA, Horwitz KB. Mapping the unique activation function 3 in the progesterone B-receptor upstream segment. Two LXXLL motifs and a tryptophan residue are required for activity. J Biol Chem. 2001;276:39843–39851. [Abstract] [Google Scholar]
- Umesono K, Evans RM. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. [Abstract] [Google Scholar]
- Vegeto E, Allan GF, Schrader WT, Tsai MJ, McDonnell DP, O’Malley BW. The mechanism of RU486 antagonism is dependent on the conformation of the carboxy-terminal tail of the human progesterone receptor. Cell. 1992;69:703–713. [Abstract] [Google Scholar]
- Vegeto E, Shahbaz MM, Wen DX, Goldman ME, O’Malley BW, McDonnell DP. Human progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B function. Mol Endocrinol. 1993;7:1244–1255. [Abstract] [Google Scholar]
- Verrijdt G, Haelens A, Schoenmakers E, Rombauts W, Claessens F. Comparative analysis of the influence of the high-mobility group box 1 protein on DNA binding and transcriptional activation by the androgen, glucocorticoid, progesterone and mineralocorticoid receptors. Biochem J. 2002;361:97–103. [Europe PMC free article] [Abstract] [Google Scholar]
- Ward RD, Weigel NL. Steroid receptor phosphorylation: Assigning function to site-specific phosphorylation. Biofactors. 2009;35:528–536. [Europe PMC free article] [Abstract] [Google Scholar]
- Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol Cell Biol. 2002;22:5451–5466. [Europe PMC free article] [Abstract] [Google Scholar]
- Wardell SE, Kwok SC, Sherman L, Hodges RS, Edwards DP. Regulation of the amino-terminal transcription activation domain of progesterone receptor by a cofactor-induced protein folding mechanism. Mol Cell Biol. 2005;25:8792–8808. [Europe PMC free article] [Abstract] [Google Scholar]
- Wardell SE, Narayanan R, Weigel NL, Edwards DP. Partial agonist activity of the progesterone receptor antagonist RU486 mediated by an amino-terminal domain coactivator and phosphorylation of serine400. Mol Endocrinol. 2010;24:335–345. [Europe PMC free article] [Abstract] [Google Scholar]
- Warnmark A, Wikstrom A, Wright AP, Gustafsson JA, Hard T. The N-terminal regions of estrogen receptor alpha and beta are unstructured in vitro and show different TBP binding properties. J Biol Chem. 2001;276:45939–45944. [Abstract] [Google Scholar]
- Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen MP, Chen D, Huang SM, Subramanian S, McKinerney E, et al. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. [Abstract] [Google Scholar]
- Weigel NL, Beck CA, Estes PA, Prendergast P, Altmann M, Christensen K, Edwards DP. Ligands induce conformational changes in the carboxyl-terminus of progesterone receptors which are detected by a site-directed antipeptide monoclonal antibody. Mol Endocrinol. 1992;6:1585–1597. [Abstract] [Google Scholar]
- Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. [Abstract] [Google Scholar]
- Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–1126. [Abstract] [Google Scholar]
- Xu J, Nawaz Z, Tsai SY, Tsai MJ, O’Malley BW. The extreme C terminus of progesterone receptor contains a transcriptional repressor domain that functions through a putative corepressor. Proc Natl Acad Sci U S A. 1996;93:12195–12199. [Europe PMC free article] [Abstract] [Google Scholar]
- Yi P, Wu RC, Sandquist J, Wong J, Tsai SY, Tsai MJ, Means AR, O’Malley BW. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1) Mol Cell Biol. 2005;25:9687–9699. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao Q, Chasse SA, Devarakonda S, Sierk ML, Ahvazi B, Rastinejad F. Structural basis of RXR-DNA interactions. J Mol Biol. 2000;296:509–520. [Abstract] [Google Scholar]
- Zhao Q, Khorasanizadeh S, Miyoshi Y, Lazar MA, Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell. 1998;1:849–861. [Abstract] [Google Scholar]
- Zilliacus J, Wright AP, Carlstedt-Duke J, Gustafsson JA. Structural determinants of DNA-binding specificity by steroid receptors. Mol Endocrinol. 1995;9:389–400. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.mce.2011.07.017
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4437577?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Unraveling the Dynamics of Estrogen and Progesterone Signaling in the Endometrium: An Overview.
Cells, 13(15):1236, 23 Jul 2024
Cited by: 1 article | PMID: 39120268 | PMCID: PMC11312103
Review Free full text in Europe PMC
The diagnostic potentiality of the RNA aptamer against progesterone receptor isolated by crush and soak (CRUSOAK)-SELEX.
Mikrochim Acta, 191(6):346, 27 May 2024
Cited by: 0 articles | PMID: 38802696
Novel biosensor for high-throughput detection of progesterone receptor-interacting endocrine disruptors.
Sci Rep, 14(1):5567, 06 Mar 2024
Cited by: 1 article | PMID: 38448539 | PMCID: PMC10917811
Differential Gene Regulation of the Human Blastocyst Trophectoderm and Inner Cell Mass by Progesterone.
Reprod Sci, 31(5):1363-1372, 27 Dec 2023
Cited by: 0 articles | PMID: 38151652
Reevaluating the Role of Progesterone in Ovarian Cancer: Is Progesterone Always Protective?
Endocr Rev, 44(6):1029-1046, 01 Nov 2023
Cited by: 2 articles | PMID: 37261958 | PMCID: PMC11048595
Review Free full text in Europe PMC
Go to all (49) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements.
Mol Endocrinol, 20(12):3042-3052, 24 Aug 2006
Cited by: 47 articles | PMID: 16931575 | PMCID: PMC2532839
Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding.
Nucleic Acids Res, 36(11):3655-3666, 12 May 2008
Cited by: 21 articles | PMID: 18474528 | PMCID: PMC2441811
Hinge and amino-terminal sequences contribute to solution dimerization of human progesterone receptor.
Mol Endocrinol, 11(8):1114-1128, 01 Jul 1997
Cited by: 43 articles | PMID: 9212059
Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations.
Biochem J, 391(pt 3):449-464, 01 Nov 2005
Cited by: 98 articles | PMID: 16238547 | PMCID: PMC1276946
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA046938
Grant ID: CA46938
NIGMS NIH HHS (5)
Grant ID: R01 GM079154-02
Grant ID: R01 GM079154-04
Grant ID: R01 GM079154
Grant ID: R01 GM079154-01A1
Grant ID: R01 GM079154-03




