Abstract
Free full text

Reciprocal functional pseudotyping of HIV-1 and HTLV-1 viral genomes by the heterologous counterpart envelope proteins
Abstract
HIV-1 and HTLV-1 can infect CD4+ T cells and can co-infect the same individual. In principle, it is possible that both viruses can infect the same CD4+ T cells in dually infected persons. Currently, how efficiently HTLV-1 and HIV-1 co-infects the same cell and the full extent of their biological interactions are not well-understood. Here, we report evidence confirming that both viruses can infect the same cells and that HTLV-1 envelope (Env) can pseudotype HIV-1 viral particles and HIV-1 envelope (Env) can pseudotype HTLV-1 virions to mediate subsequent infections of substrate cells. We also show that the construction of a chimeric HTLV-1 molecular clone carrying the HIV-1 Env in place of its HTLV-1 counterpart results in a replication competent moiety. These findings raise new implications of viral complementation and assortment between HIV-1 and HTLV-1 in dually infected persons.
INTRODUCTION
Human T-cell Leukemia Virus (HTLV-1) and Human Immunodeficiency Virus (HIV-1) are the two known pathogenic human retroviruses (Gallo, 2005). Worldwide, approximately 20 million people are infected with HTLV-1 (Proietti et al., 2005). Infection by HTLV-1 is associated with the development of Adult T-cell Leukemia (ATL) in a minority of individuals following a lengthy latency period (20–40 years). HTLV-1 infection with is also associated with inflammatory diseases such as HTLV-1 Associated Myelopathy / Tropical Spastic Paraparesis (HAM/TSP) (Gessain and Mahieux, 2012).
HIV-1 infection leads to an acute phase of infection lasting several weeks that is characterized by a high plasma viral load. Following the acute period, levels of HIV-1 in the blood drop to a set point that characterizes chronic infection (Fauci et al., 1996). Over time the infected person’s CD4+ T-cell count decreases and, in the absence of therapy, most individuals acquire opportunistic infections that usually progress to death (Fauci et al., 1996). The HIV-1 envelope (Env) binds the CD4 receptor (Wilen et al., 2012), while the receptor for HTLV-1 remains controversial (Ghez et al., 2006; Jin et al., 2006; Manel et al., 2003; Pinon et al., 2003). However, the virus is known to be capable of infecting CD4+ and CD8+ T cells (Eiraku et al., 1998; Jones et al., 2008; Manel et al., 2005).
HIV-1 infected individuals can be co-infected with a variety of other viruses. Several studies have reported co-infection of HIV-1 with herpesviruses (da Silva and de Oliveira, 2011), hepatitis B virus (HBV) (Martin-Carbonero and Poveda, 2012), hepatitis C virus (HCV) (Taylor et al., 2012) and other viruses. In each of these examples, while the host is infected with multiple viruses, individual cells are infected with only one virus due to tropism. Thus, in such co-infections, the different viral genomes and their gene products are unlikely to co-exist or interact within the same cells.
HIV-1 and HTLV-1 co-infection in vivo has also been extensively reported, and it has been suggested that co-infection by the two viruses influences the pathologic progression of both viruses (Bahia et al., 2011; Beilke, 2012; Brites et al., 2009; Pedroso et al., 2011). Because both HIV-1 and HTLV-1 can infect CD4+ T cells, this co-infection may differ from those of HIV-1/herpesviruses in that both viral genomes could co-exist in a single infected cell. Although several labs have examined co-infection at the cellular level (Montefiori and Mitchell, 1987; Spear et al., 1991; Szabo et al., 1999), these analyses report either very rare cases of co-infection or rely on the developmental of a cell line stably infected with either HIV-1 or HTLV-1. A clone of this stable infection is then expanded and infected with the other virus. Herein, we re-examined the issue and demonstrated de novo co-infection of the same cell using flow cytometry. We found that co-infection can lead to psuedotyping of the HIV-1 virion with HTLV-1 Env. To examine the reciprocal nature of pseudotyping, we also created a chimeric HTLV-1 that carries the HIV-1, but not the HTLV-1, Env. We demonstrated that this virus can competently replicate and compete with WT HIV-1 in a spreading viral replication.
Super infection exclusion describes the property of virally infected cells to be resistant to re-infection (super infection) with a new incoming virion (Nethe et al., 2005; Potash and Volsky, 1998). (1;2) Super infection exclusion has been described for a variety of human viral diseases including; influenza A, Rubella, hepatitis C, foamy virus, rhabdoviruses, HSV-1, vaccinia, flaviviruses and HIV-1 (Berg et al., 2003; Bock et al., 1998; Claus et al., 2007; Giannecchini et al., 2007; Lee et al., 2005; Mador et al., 2002; Nethe et al., 2005; Tscherne et al., 2007; Turner and Moyer, 2008; Whitaker-Dowling et al., 1983). In HIV-1 infection Nef, Env and Vpu down regulate cell surface expression of the CD4 receptor molecule (Nethe et al., 2005; Wildum et al., 2006). Binding of Nef to CD4 and a set of multiple adapter proteins leads to endocytosis and degradation of CD4 (Lindwasser et al., 2007; Rose et al., 2005). In contrast to Nef, Env and Vpu work to prevent transport of newly synthesized CD4 molecules to the cell surface (Lindwasser et al., 2007; Wildum et al., 2006). The ability of our HTLV-1 chimera to inhibit HIV-1 replication suggests that Env alone may be sufficient to mediate super-infection exclusion.
RESULTS
HTLV-1 and HIV-1 infect the same cell
To examine HTLV-1 and HIV-1 infection of the same cell, we employed the J-LTR-G reporter cell line (Kutsch et al., 2004) and antibody specific to HTLV-1 Env gp46. J-LTR-G is a Jurkat T-cell line that carries an HIV-1 LTR driving a GFP reporter; GFP is produced when this cell line is infected with HIV-1. For our experiments, J-LTR-G cells were cultured alone (Fig. 1A, first panel), with HIV-1 NL4-3 (Fig. 1A, second panel), with HTLV-1 transformed virus-producer cell line MT2 (Fig. 1A, third panel), or with HTLV-1 transformed virus-producer cell line MT2 and HIV-1 NL4-3 (Fig. 1A, fourth panel). Uninfected J-LTR-G cells showed background staining for gp46 and a background level of GFP (Fig. 1A, first panel). Co-culture of J-LTR-G with MT2 resulted in the detection of cells positively stained for HTLV-1 gp46 (Fig. 1, second panel). The flow cytometric profile shows two populations with the MT2 cells distinguishable as a brightly stained smaller discrete cell population (red arrow) while the gp46 positive J-LTR-G cells are represented by a rightward shifted larger cell population (black arrow). The rightward-shift of the J-LTR-G population along the gp46 staining axis is consistent with HTLV-1 virions produced from MT2 cells successfully infecting the former cells. Infection of J-LTR-G with HIV-1 NL4-3 produced, as expected, a population of GFP positive cells (12.6%; Fig. 1A third panel), and the co-culture of J-LTR-G with MT2 and HIV-1 NL4-3 produced cell populations that are positive for both gp46 and GFP (39.7%; Fig. 1A fourth panel). The latter cells (39.7% GFP+gp46+) represent cells that are simultaneously infected with both HTLV-1 and HIV-1. An interesting observation from this experiment was that co-infection with HTLV-1 increased the number of GFP (HIV-1) positive cells. To insure that this was not an artifact, a follow-up experiment was performed in the setting of spreading infection that also indicated that infection with HTLV-1 could increase HIV-1 replication (Supplemental Figure S1).

A) Jurkat indicator cells (J-LTR-G) were infected with NL4-3 HIV-1 viral stocks. Four days post HIV-1 infection the cells were co-cultured 10:1 with HTLV-1 infected cell line MT2. After three days of co-culture infection was detected by flow cytometry for GFP (HIV-1) or cell surface gp46 (HTLV-1) using an AlexaFluor547 conjugated anti-gp46. Black arrow indicates HTLV infected J-LTR-G cells, while the red arrow indicates the MT-2 cells from co-culture. B) Jurkat cells were placed in the bottom well of a transwell plate. ACH2 and MT2 cells were stimulated with 1 µm PMA for twenty-four hours, washed and placed in the top of the transwell system. Seventy-two hours later the Jurkat cells were removed, affixed to a microscope slide and stained for HIV-1 gp120 (Red), HTLV-1 Tax (Green) or DNA by DAPI (Blue). Yellow color in the merged panel indicates co-staining for both proteins.
To confirm the above results in a different system we chose to examine co-infection by fluorescent microscopy. In brief, Jurkat T-cells were placed in the bottom section of a transwell tissue culture plate. PMA stimulated ACH2 and MT-2 cells were placed in the upper well and the cultured carried for 72 hours. Jurkat T-cells were then affixed to a slide and stained for HTLV-1 Tax or HIV-1 Envelope (Figure 1B). This staining confirmed the results of the flow cytometry, in that uninfected, singly infected and doubly infected cells were all found in the same culture.
Co-infection generates HIV-1 virions pseudotyped by the HTLV-1 Env
We next asked if co-infection of a cell by HIV-1 and HTLV-1 could lead to pseudotyping of the former by the latter envelope protein. Because an HIV-1 virion carrying an HTLV-1 envelope could allow the broader HIV-1 tropism, we first examined HIV-1 pseudotyping by the HTLV-1 envelope (Fig 2). Accordingly, we prepared VSV-G pseudotyped HIV-1 particles by cotransfecting a VSV-G expression vector and an envelope-deleted NL4-3 ΔEnv GFP (Zhang et al., 2004) molecular clone into 293T cells. Supernatant containing VSV-G pseudotyped HIV-1 virions was harvested from the 293T cells 48 hours after transfection and filtered; the filtrate was used to infect either Jurkat or MT2 cells. Three days later, the infected Jurkat or MT2 cells were separately co-cultured with the HIV-1 indicator cell line TZMbl. We expected that the envelope-deleted NL4-3 ΔEnv GFP genome would not produce infectious particles in Jurkat cells because these cells express no retroviral Env protein. On the other hand because MT2 cells do express HTLV-1 Env protein (Harada et al., 1985). Should this envelope protein be competent for pseudotyping HIV-1, then HTLV-1 Env pseudotyped HIV-1 particles should be produced. In that setting, HTLV-1 Env pseudotyped HIV-1 virions could be detected by an infection assay using TZMbl cells. Indeed, β-galactosidase staining of a co-culture of NL4-3 ΔEnv GFP/MT2 with TZMbl cells shows positive staining. The control co-culture of NL4-3 ΔEnv GFP/Jurkat with TZMbl does not (Fig 2A, B).

Jurkat or MT2 cells were infected with mock supernatant or NL4-3 ΔEnv pseudotyped with VSV-G. Three days post infection, cells were co-cultured with the HIV-1 indicator cell line TZMbl. One day after co-culture, infection with HIV-1 was detected by b β-galactosidase staining of the TZMbl cells. A) Representative micrographs of the four conditions. B) Triplicate experiments were performed to determine the number of β-galactosidase positive TZMbl cells following co-culture. C) Infection of MT2 cells with mock or NL4-3 ΔEnv pseudotyped with VSV-G was performed as above. Co-culture was performed in the presence of increasing concentrations of anti-anti-HIV-1 (VRC01) or anti-HTLV-1 neutralizing antibodies. ** p-value ≤ 0.01, *** p-value ≤ 0.001
To further verify that the results from co-culture of NL4-3 ΔEnv GFP/MT2 with TZMbl cells were due to pseudotyping of NL4-3 ΔEnv GFP with the HTLV-1 Env from MT2, we repeated the co-culturing assay in the presence of neutralizing antibodies specific for HIV-1 or HTLV-1 envelope proteins (Fig 2C). In the presence of anti-HTLV-1 antibody, a dose-dependent inhibition of TZMbl reporter readout was seen, while in the presence of anti-HIV-1 antibody a small change could be observed only at the highest antibody concentration. These results support the interpretation that infectious NL4-3 ΔEnv GFP virions arise after pseudotyping by HTLV-1 Env protein.
Construction of a chimeric HTLV-1 genome that expresses HIV-1 Envelope
We took a different approach to investigate whether HTLV-1 could be pseudotyped with HIV-1 Env. Using the HTLV-1 ACH molecular clone (Kimata et al., 1994), we created a chimeric HTLV-1 genome in which the native Env gene was replaced with the HIV-1 Env gene from the NL4-3 molecular clone (Fig. 3). We reasoned that if HIV-1 Env could complement HTLV-1 Env function, then this chimeric HTLV-1 genome should be replication competent. In our chimera, we modeled our cloning strategy after that performed by Delebecque et al. in which the HTLV-I Env in a molecular clone was replaced by a heterologous Env gene from MuMLV (Delebecque et al., 2005; Delebecque et al., 2002). In order not to perturb the splicing and processing of the HTLV-I genome, the signal peptide from the HTLV-1 Env gene was fused to the NL4-3 HIV-1 Env resulting in a modified Env whereby the signal peptide of HIV-1 Env was replaced with the HTLV-I signal peptide. This chimeric Env was then inserted back into the ACH molecular clone using the SphI and NsiI restriction sites (Fig 3A). Although this construct will not express HIV-1 Rev, the HTLV-1 Rex coding sequence remains un-perturbed. Rex has been shown to be capable of stabilizing HIV-1 Env, complementing Rev function and will allow expression in this chimera (Hanly et al., 1989).

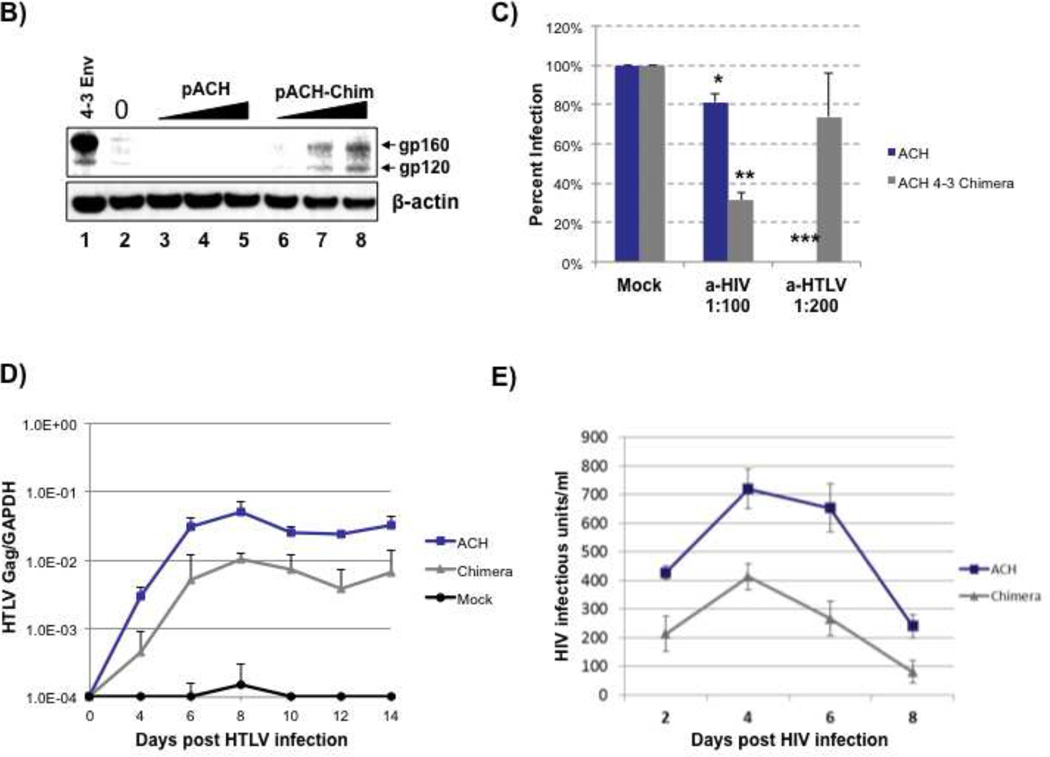
A) Signal peptide from HTLV-I Env (top panel) was fused to the HIV-1 Env sequence (middle panel) to create a chimeric Env (bottom panel) which was then transferred back into the pACH molecular clone by way of the SphI and NsiI sites. This process results in the preservation of HTLV-I splicing sequences and perturbs no viral genes apart from the Env. B) Western blot analysis of expression of HIV-1 Env after transfection of seeded 2×105 TZMbl cells with 0, 0.25, 0.5 or 1 microgram of pACH or pACH-Chimera. Arrows indicate gp160 and gp120. Extracts were used to blot for β-actin to insure equal loading. C) 5×104 Jurkat T-cells were seeded in each well of a 96 well plate and infected with ACH or ACH-Chimera in the presence of αHIV-1 nAb (1:100) or αHTLV-1 nAb (1:200). Eight days post-infection RNA was isolated from the cells and used to determine relative infection by qPCR for HTLV-1 Gag. * p-value ≤ 0.05, ** p-value ≤ 0.01, *** p-value ≤ 0.001 D) 6×106 Jurkat T-cells were infected with equal amounts of ACH or ACH-Chimera molecular clones as determined by p19 Elisa. RNA was extracted from infected cells at days 4, 6, 8, 10, 12 and 14 post infection. Quantitative RT-PCR was used to determine the expression of HTLV-1 Gag mRNA as compared to cellular GAPDH mRNA. E) ACH or Chimera infected Jurkat T-cells from day 4 of infection were exposed to equal quantities of NL4-3 HIV-1 molecular clone. Supernatant was harvested at days 2, 4, 6 and 8 post infection. HIV-1 replication was determined by measuring infectious units per ml of culture media in TZMbl cells.
We next determined if the pACH-Chimera proviral genome could express HIV-1 Env. TZMbl cells were transfected separately with increasing amounts of pACH or pACH-Chimera, or with a pCMV 4-3 Env expression vector as control (Fig 3B). Western blotting using human HIV-1 hyper-immune serum showed that increasing the transfection of pACH-Chimera led to increasing expression of Env protein in the TZMbl cells (Fig. 3B, lanes 6–8) which co-migrated with envelope protein produced from transfected pCMV 4-3 Env expression vector (Fig. 3B, lane 1). In contrast the transfection of the parental pACH genome produced no HIV-1 Env protein (Fig. 3B, lanes 3–5).
To test the infectivity of our ACH-Chimera virus, we generated viral stocks by transfecting 293T cells with 10 µg of pACH with 1 µg pCTax (CMV promoter-driven Tax) or 10 µg pACH-Chimera with 1 µg pCTax. Seventy-two hours after transfection supernatants were harvested, filtered, and HTLV-1 virus production was determined by HTLV-1 p19 Gag Elisa (Zeptometrix, Buffalo, NY). To characterize the ACH and ACH-Chimera viruses, we asked if we could distinguish between the two viruses based on their envelope proteins. Thus, we challenged ACH-virus or ACH-Chimera particles with either HIV-1 or HTLV-1 neutralizing antibodies (Fig 3C). Accordingly, Jurkat T-cells were infected with p19 Gag-equivalent amounts of either ACH WT or ACH Chimera in the presence of neutralizing antibodies specific for HIV-1 or HTLV-1. Eight days post infection the level of cell associated viral RNA was determined by quantitative RT-PCR of total cellular RNA, and we observed that the ACH-virus was sensitive to HTLV-1 neutralizing antibody while the ACH-Chimera was sensitive to HIV-1 neutralizing antibody, consistent with the expression and function of HIV-1 Env expressed from the latter genome (Fig. 3C).
We next determined the ability of the ACH-Chimera virus to replicate in tissue culture. Jurkat T-cells were infected with ACH or ACH-Chimera virus, normalized by HTLV-1 p19 Gag measurements. RNA was extracted from infected cells at 4, 6, 8, 10, 12 and 14 days post infection, and HTLV-1 viral RNA was quantified by q-RT-PCR (Fig 3D). The results showed that both ACH and ACH-Chimera were replication competent with the latter replicating approximately 0.5 to 1 log less well than the wild type ACH.
ACH-Chimera suppresses HIV-1 replication
Previous findings suggest that homologous Env expression may sufficiently mediate virus super infection exclusion. As an additional functional test, we asked if infection of Jurkat T-cells by ACH-Chimera could elicit super infection exclusion of wild type HIV-1. Jurkat T-cells were infected with equivalent amounts of ACH or ACH-Chimera, as determined by p19 elisa. Four days after infection, the cells were challenged with a second infection using HIV-1 NL4-3. Supernatant was collected and assayed for the presence of infectious HIV-1 particles by exposing the TZMbl indicator cell line to culture supernatants and assaying for Tat induced β-galactosidase expression (Figure 3E). The read out from a Tat responsive reporter allowed us to accurately measure and segregate HIV-1 from HTLV-1 production. In repeated assays, prior infection with ACH-Chimera compared to ACH consistently reduced production of HIV-1 via a subsequent super infection. This result is consistent with functional HIV-1 Env expression from ACH-Chimera excluding super infection of the cells by HIV-1 NL4-3.
We were intrigued that Env alone could be effective at blocking infection. Env is thought to work in concert with Vpu and Nef to down-regulate CD4 to block infection (Lindwasser et al., 2007; Wildum et al., 2006). However, CD4 down-regulation occurs late in infection, often just before cell death, and may be more important for viral release (Arganaraz et al., 2003; Benson et al., 1993; Nethe et al., 2005). Further, there is evidence that resistance to super infection occurs early in infection and when CD4 levels are still high (Saha et al., 1999; Volsky et al., 1996). This led us to posit that Env might be blocking infection without altering CD4 expression; possibly by binding CD4 at the cell surface. To test this hypothesis we transfected TZMbl with a GFP expression vector and 0.5 or 1 µg of Env expression vector. Twenty-four hours post transfection the cells were infected with NL4-3 at an MOI of 0.2. Twenty-four hours post infection the cells were imaged for GFP and β-galactosidase expression. The number of β-gal (HIV-1 infected) cells per GFP positive (transfected cells) was then determined (Supplemental Fig. S2A). Transfection of cells with wild-type (WT) or fusion-peptide mutant (FP Mut) Env was capable of reducing infection by NL4-3. However, a mutant incapable of binding CD4 (BS Mut) did not mediate this effect. Further, no down-regulation of CD4 was detected after transfection with wild-type Env (Supplemental Fig. S2B).
CONCLUSIONS
Although there have been clear reports of patients co-infected with both HIV-1 and HTLV-1, evidence for co-infection at the cellular level is unclear (Bahia et al., 2011; Beilke, 2012; Pedroso et al., 2011). Herein, we demonstrate de novo infection of HTLV-I producing cells (MT2) by cell free infection with HIV-1. We further demonstrate that co-infection of MT2 cells lead to pseudotyping of HIV-1 with HTLV-1 envelope. Because HTLV-1 Env is thought to have a broader tropism than HIV-1 Env, this finding suggests that such pseudotyping could possibly lead to single round infection of non-classical target cells by HIV-1. Nevertheless, the biological relevance of this observation to HIV-1 pathogenesis in dually co-infected hosts remains unknown.
We next determined whether the reciprocal finding could be shown (i.e. pseudotyping of HTLV-1 by HIV-1 Env). We created a chimeric HTLV-1 virus that substituted the HIV-1 Env for its own envelope protein. This virus expresses the HIV-1 envelope and is competent for a spreading infection in a T-cell line (Fig 3). Furthermore, infection with the chimeric HTLV-1 (ACH-Chimera) virus altered the replication of a subsequent HIV-1 infection. The above observation suggests that envelope expression-alone is sufficient to mediate super infection exclusion. It raises the possibility that pathologically attenuated HTLV-1 vector that replicates using an HIV-1 Env could be a gene-based strategy against HIV-1 infection.
METHODS
Cell culture and transfections
Adherent cell lines (TZMbl and 293T) were maintained in DMEM supplemented with 10% fetal bovine serum, L-glutamine and penicillin/streptomycin. Adherent cell lines were transfected using Lipofectamine LTX (Invitrogen) according the manufacturer’s protocol. Suspension cell lines (Jurkat, J-LTR-G and MT2) were maintained in RPMI supplemented with 10% fetal bovine serum, L-glutamine and penicillin/streptomycin.
Flow cytometry
Infection of J-LTR-G cells was followed by flow cytometry. Cells were stained using anti-gp46 antibody (Abcam) for one hour, washed and then stained with anti-mouse AlexaFluor647 (Invitrogen). Staining for cell surface gp46 and GFP expression was detected using a Becton Dickinson Fortessa LSR.
Fluorescent Microscopy
Jurkat cells were affixed to a positively charged glass slide through use of a CytoSpin and fixed with 1% paraformaldehyde for 10 minutes. Cells were then washed, blocked in PBS with 1% BSA and 0.1% NP40 and stained with goat anti-gp120 (1:500) (AbCam) or mouse anti-Tax hybridoma supernatant (1:200). After another series of washes, the cells were stained with anti-goat antibody conjugated to AlexaFluor594 (1:1000) (Invitrogen) and anti-mouse AlexaFluor488 (1:1000) (Invitrogen). Cells were then washed, stained with DAPI and imaged use a fluorescent microscope.
Infection and neutralization assays
TZMbl cells were used to determine infection via HIV-1 (either WT or pseudotyped). Cells were incubated with cell free virus or co-cultured with source cells for twenty-four hours. After incubation with virus, the cells were fixed and stained for expression of β-galactosidase fluorescently using the ImaGeneRed LacZ kit (Invitrogen). For neutralization assays, cells were incubated with anti-HIV-1 VRC01 or anti-HTLV-1 antiserum (AIDS Reagent Program) concomitantly with infecting virus. Final dilutions of antibody were 1:100, 1:200 or 1:1000 as indicated.
Viral infections
HIV-1 viral stocks were generated by transfecting the proviral plasmid pNL4-into 293T cells. Seventy-two hours after transfection, the culture supernatant was clarified by centrifugation and filtered using a 0.22 μM filter to remove remaining cells. Viral stocks were quantified using reverse transcriptase assay and titered on TZMbl cells. Infection of TZMbl cells was determined by β-galactosidase staining and replication in Jurkat T-cells was followed by reverse transcriptase assay.
HTLV-1 viral stocks were generated by transfecting 293T with pACH proviral plasmid and pCMV-Tax at a ratio of 10:1. Seventy-two hours after transfection the culture supernatant was clarified by centrifugation, filtered using a 0.22 μM filter to remove remaining cells and concentrated by ultracentrifugation. Viral stocks were quantified using p19 Elisa (Zeptometrix). For cell free infection, 6×106 Jurkat cells were incubated with HTLV-1 viral stock containing 10ng p19 in 2 ml media for 16 hours. Infected cells were then washed and re-suspended in media at 1×106 cells/ml. Infection was followed by extracting cell associated RNA from the cells every other day and virus replication was detected by qPCR for HTLV-1 Gag
Construction of chimeric HTLV-1
The leader peptide of HTLV-1 Env was PCR amplified from the X1MT HTLV-1 molecular clone using primers X1MT 5226 F and HTLV Env ExtR. The HTLV Env Ext R primer contains sequence matching the desired junction with the HIV-1 envelope and the KpnI site found in the HIV-1 Envelope. Amplified HTLV-1 leader peptide plus adapter was digested with SphI and KpnI and ligated into the SphI and KpnI sites of pUC19 to created pUC-3’HTLVEnv.
A fragment of the HIV-1 Env was amplified from pNL4-3 using HIV Env F and HIV 8785 EcoRI R primers. The resulting PCR fragment was digested with KpnI and EcoRI and ligated into the KpnI and EcoRI sites of pUC-3’HTLVEnv. The complete envelope containing fragment was then amplified by PCR using primers Chim PstI and Chim R (below). Fragment was digested using SphI and PstI and ligated into pACH using the SphI and NsiI sites.
Primers
| HTLV Gag For | CCTTCGTAGAACGCCTCAAC |
| HTLV Gag Rev | CAAGCCCGCAACATATCTCC |
| X1MT 5226 F | ATAGCCGCCAGTGGAAAGGACC |
| HTLV Env Ext R | CAGGTACCCCATAATAGACTGTGACCCACAATTTTTCTGTACCGAGGATGAGGGGGCAG |
| HIV Env F | CTGCCCCCTCATCCTCGGTACAGAAAAATTGTGGGTCAC |
| Chim PstI | CTGCAGAAAACGACGGCCAGTG |
| Chim R | ATGCTTCCGGCTCGTATG |
Supplementary Material
01
Supplemental Figure 1: HTLV-1 infection leads to greater replication of HIV-1:ACH or mock infected Jurkat T-cells from day 4 of infection were exposed to equal quantities of NL4-3 HIV-1 molecular clone. Supernatant was harvested at days 2, 4, 6 and 8 post infection. HIV-1 replication was determined by measuring infectious units per ml of culture media in TZMbl cells.
02
Supplemental Figure 2: Env mediates super-infection resistance independent of CD4d own regulation:A) TZMbl cells were seeded 2×105 cells/well in a 12-well tissue culture plate and transfected with 0, 0.5 or 1 microgram of pCMV 4-3-Env (WT) pCMV 4-3-BS (BS Mut) or pCMV 4-3-FP (FP Mut) and 1 microgram pMAX-GFP. DNA level was held constant with an un-related CMV expression vector. Cells were then infected 24 hours later with NL4-3. Cultures were fixed twenty-four hours after infection and stained using Imagene Red lacZ (Invitrogen) and imaged. Numbers indicate the relative infection as compared to control. All data is the average of three replicates. * p-value ≤ 0.05, ** p-value ≤ 0.01 B) TZMbl cells were seeded 2×105 cells/well in a 12-well tissue culture plate and transfected with 0, 0.25, 1 or 2 micrograms of pCMV 4-3-Env and 1 microgram of pMAX-GFP. Forty-eight hours post transfection cells were fixed in 1% formaldehyde and stained for CD4 expression using PE conjugated anti-CD4 antibody. GFP positive transfected cells were analyzed for cell surface expression of CD4. Histograms indicate the CD4-PE signal for the GFP positive population of each transfection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arganaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. The Journal of biological chemistry. 2003;278:33912–33919. [Abstract] [Google Scholar]
- Bahia F, Novais V, Evans J, Le Marchand C, Netto E, Page K, Brites C. The impact of human T-cell lymphotropic virus I infection on clinical and immunologic outcomes in patients coinfected with HIV and hepatitis C virus. J Acquir Immune Defic Syndr. 2011;57(Suppl 3):S202–S207. [Europe PMC free article] [Abstract] [Google Scholar]
- Beilke MA. Retroviral coinfections: HIV and HTLV: taking stock of more than a quarter century of research. AIDS research and human retroviruses. 2012;28:139–147. [Europe PMC free article] [Abstract] [Google Scholar]
- Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. The Journal of experimental medicine. 1993;177:1561–1566. [Europe PMC free article] [Abstract] [Google Scholar]
- Berg A, Pietschmann T, Rethwilm A, Lindemann D. Determinants of foamy virus envelope glycoprotein mediated resistance to superinfection. Virology. 2003;314:243–252. [Abstract] [Google Scholar]
- Bock M, Heinkelein M, Lindemann D, Rethwilm A. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology. 1998;250:194–204. [Abstract] [Google Scholar]
- Brites C, Sampalo J, Oliveira A. HIV/human T-cell lymphotropic virus coinfection revisited: impact on AIDS progression. AIDS reviews. 2009;11:8–16. [Abstract] [Google Scholar]
- Claus C, Tzeng WP, Liebert UG, Frey TK. Rubella virus-induced superinfection exclusion studied in cells with persisting replicons. The Journal of general virology. 2007;88:2769–2773. [Abstract] [Google Scholar]
- da Silva SR, de Oliveira DE. HIV, EBV and KSHV: viral cooperation in the pathogenesis of human malignancies. Cancer letters. 2011;305:175–185. [Abstract] [Google Scholar]
- Delebecque F, Combredet C, Gabet AS, Wattel E, Brahic M, Tangy F. A chimeric human T cell leukemia virus type I bearing a deltaR Moloney-murine leukemia virus envelope infects mice persistently and induces humoral and cellular immune responses. The Journal of infectious diseases. 2005;191:255–263. [Abstract] [Google Scholar]
- Delebecque F, Pramberger K, Prevost MC, Brahic M, Tangy F. A chimeric human T-cell lymphotropic virus type 1 with the envelope glycoprotein of Moloney murine leukemia virus is infectious for murine cells. Journal of virology. 2002;76:7883–7889. [Europe PMC free article] [Abstract] [Google Scholar]
- Eiraku N, Hingorani R, Ijichi S, Machigashira K, Gregersen PK, Monteiro J, Usuku K, Yashiki S, Sonoda S, Osame M, Hall WW. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotropic virus type I-infected individuals. Journal of immunology. 1998;161:6674–6680. [Abstract] [Google Scholar]
- Fauci AS, Pantaleo G, Stanley S, Weissman D. Immunopathogenic mechanisms of HIV infection. Annals of internal medicine. 1996;124:654–663. [Abstract] [Google Scholar]
- Gallo RC. History of the discoveries of the first human retroviruses: HTLV-1 and HTLV-2. Oncogene. 2005;24:5926–5930. [Abstract] [Google Scholar]
- Gessain A, Mahieux R. Tropical spastic paraparesis and HTLV-1 associated myelopathy: clinical, epidemiological, virological and therapeutic aspects. Revue neurologique. 2012;168:257–269. [Abstract] [Google Scholar]
- Ghez D, Lepelletier Y, Lambert S, Fourneau JM, Blot V, Janvier S, Arnulf B, van Endert PM, Heveker N, Pique C, Hermine O. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. Journal of virology. 2006;80:6844–6854. [Europe PMC free article] [Abstract] [Google Scholar]
- Giannecchini S, Pistello M, Isola P, Matteucci D, Mazzetti P, Freer G, Bendinelli M. Role of Env in resistance of feline immunodeficiency virus (FIV)-infected cats to superinfection by a second FIV strain as determined by using a chimeric virus. Journal of virology. 2007;81:10474–10485. [Europe PMC free article] [Abstract] [Google Scholar]
- Hanly SM, Rimsky LT, Malim MH, Kim JH, Hauber J, Duc Dodon M, Le SY, Maizel JV, Cullen BR, Greene WC. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes & development. 1989;3:1534–1544. [Abstract] [Google Scholar]
- Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. [Abstract] [Google Scholar]
- Jin Q, Agrawal L, VanHorn-Ali Z, Alkhatib G. Infection of CD4+ T lymphocytes by the human T cell leukemia virus type 1 is mediated by the glucose transporter GLUT-1: evidence using antibodies specific to the receptor's large extracellular domain. Virology. 2006;349:184–196. [Abstract] [Google Scholar]
- Jones KS, Petrow-Sadowski C, Huang YK, Bertolette DC, Ruscetti FW. Cell-free HTLV-1 infects dendritic cells leading to transmission and transformation of CD4(+) T cells. Nature medicine. 2008;14:429–436. [Abstract] [Google Scholar]
- Kimata JT, Wong FH, Wang JJ, Ratner L. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology. 1994;204:656–664. [Abstract] [Google Scholar]
- Kutsch O, Levy DN, Bates PJ, Decker J, Kosloff BR, Shaw GM, Priebe W, Benveniste EN. Bis-anthracycline antibiotics inhibit human immunodeficiency virus type 1 transcription. Antimicrobial agents and chemotherapy. 2004;48:1652–1663. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. Journal of virology. 2005;79:3231–3242. [Europe PMC free article] [Abstract] [Google Scholar]
- Lindwasser OW, Chaudhuri R, Bonifacino JS. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Current molecular medicine. 2007;7:171–184. [Abstract] [Google Scholar]
- Mador N, Panet A, Steiner I. The latency-associated gene of herpes simplex virus type 1 (HSV-1) interferes with superinfection by HSV-1. Journal of neurovirology. 2002;8(Suppl 2):97–102. [Abstract] [Google Scholar]
- Manel N, Battini JL, Taylor N, Sitbon M. HTLV-1 tropism and envelope receptor. Oncogene. 2005;24:6016–6025. [Abstract] [Google Scholar]
- Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell. 2003;115:449–459. [Abstract] [Google Scholar]
- Martin-Carbonero L, Poveda E. Hepatitis B virus and HIV infection. Seminars in liver disease. 2012;32:114–119. [Abstract] [Google Scholar]
- Montefiori DC, Mitchell WM. Persistent coinfection of T lymphocytes with HTLV-II and HIV and the role of syncytium formation in HIV-induced cytopathic effect. Virology. 1987;160:372–378. [Abstract] [Google Scholar]
- Nethe M, Berkhout B, van der Kuyl AC. Retroviral superinfection resistance. Retrovirology. 2005;2:52. [Europe PMC free article] [Abstract] [Google Scholar]
- Pedroso C, Netto EM, Weyll N, Brites C. Coinfection by HIV-1 and human lymphotropic virus type 1 in Brazilian children is strongly associated with a shorter survival time. J Acquir Immune Defic Syndr. 2011;57(Suppl 3):S208–S211. [Abstract] [Google Scholar]
- Pinon JD, Klasse PJ, Jassal SR, Welson S, Weber J, Brighty DW, Sattentau QJ. Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans. Journal of virology. 2003;77:9922–9930. [Europe PMC free article] [Abstract] [Google Scholar]
- Potash MJ, Volsky DJ. Viral interference in HIV-1 infected cells. Reviews in medical virology. 1998;8:203–211. [Abstract] [Google Scholar]
- Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. Global epidemiology of HTLV-I infection and associated diseases. Oncogene. 2005;24:6058–6068. [Abstract] [Google Scholar]
- Rose JJ, Janvier K, Chandrasekhar S, Sekaly RP, Bonifacino JS, Venkatesan S. CD4 down-regulation by HIV-1 and simian immunodeficiency virus (SIV) Nef proteins involves both internalization and intracellular retention mechanisms. The Journal of biological chemistry. 2005;280:7413–7426. [Abstract] [Google Scholar]
- Saha K, Volsky DJ, Matczak E. Resistance against syncytium-inducing human immunodeficiency virus type 1 (HIV-1) in selected CD4(+) T cells from an HIV-1-infected nonprogressor: evidence of a novel pathway of resistance mediated by a soluble factoRs) that acts after virus entry. Journal of virology. 1999;73:7891–7898. [Europe PMC free article] [Abstract] [Google Scholar]
- Spear GT, Jiang HX, Sullivan BL, Gewurz H, Landay AL, Lint TF. Direct binding of complement component C1q to human immunodeficiency virus (HIV) and human T lymphotrophic virus-I (HTLV-I) coinfected cells. AIDS research and human retroviruses. 1991;7:579–585. [Abstract] [Google Scholar]
- Szabo J, Beck Z, Csoman E, Liu X, Andriko I, Kiss J, Bacsi A, Ebbesen P, Toth FD. Differential patterns of interaction between HIV type 1 and HTLV type I in monocyte-derived macrophages cultured in vitro: implications for in vivo coinfection with HIV type 1 and HTLV type. IAIDS research and human retroviruses. 1999;15:1653–1666. [Abstract] [Google Scholar]
- Taylor LE, Swan T, Mayer KH. HIV coinfection with hepatitis C virus: evolving epidemiology and treatment paradigms. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;55(Suppl 1):S33–S42. [Europe PMC free article] [Abstract] [Google Scholar]
- Tscherne DM, Evans MJ, von Hahn T, Jones CT, Stamataki Z, McKeating JA, Lindenbach BD, Rice CM. Superinfection exclusion in cells infected with hepatitis C virus. Journal of virology. 2007;81:3693–3703. [Europe PMC free article] [Abstract] [Google Scholar]
- Turner PC, Moyer RW. The vaccinia virus fusion inhibitor proteins SPI-3 (K2) and HA (A56) expressed by infected cells reduce the entry of superinfecting virus. Virology. 2008;380:226–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Volsky DJ, Simm M, Shahabuddin M, Li G, Chao W, Potash MJ. Interference to human immunodeficiency virus type 1 infection in the absence of downmodulation of the principal virus receptor, CD4. Journal of virology. 1996;70:3823–3833. [Europe PMC free article] [Abstract] [Google Scholar]
- Whitaker-Dowling P, Youngner JS, Widnell CC, Wilcox DK. Superinfection exclusion by vesicular stomatitis virus. Virology. 1983;131:137–143. [Abstract] [Google Scholar]
- Wildum S, Schindler M, Munch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. Journal of virology. 2006;80:8047–8059. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilen CB, Tilton JC, Doms RW. Molecular mechanisms of HIV entry. Advances in experimental medicine and biology. 2012;726:223–242. [Abstract] [Google Scholar]
- Zhang H, Zhou Y, Alcock C, Kiefer T, Monie D, Siliciano J, Li Q, Pham P, Cofrancesco J, Persaud D, Siliciano RF. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. Journal of virology. 2004;78:1718–1729. [Europe PMC free article] [Abstract] [Google Scholar]
Citations & impact
Impact metrics
Citations of article over time
Article citations
A complex remodeling of cellular homeostasis distinguishes RSV/SARS-CoV-2 co-infected A549-hACE2 expressing cell lines.
Microb Cell, 11:353-367, 08 Oct 2024
Cited by: 0 articles | PMID: 39421150 | PMCID: PMC11486504
Co-Infections and Superinfections between HIV-1 and Other Human Viruses at the Cellular Level.
Pathogens, 13(5):349, 24 Apr 2024
Cited by: 0 articles | PMID: 38787201 | PMCID: PMC11124504
Review Free full text in Europe PMC
Pseudotyping of HIV-1 with Human T-Lymphotropic Virus 1 (HTLV-1) Envelope Glycoprotein during HIV-1-HTLV-1 Coinfection Facilitates Direct HIV-1 Infection of Female Genital Epithelial Cells: Implications for Sexual Transmission of HIV-1.
mSphere, 3(2):e00038-18, 04 Apr 2018
Cited by: 4 articles | PMID: 29624497 | PMCID: PMC5885023
HTLV-1, the Other Pathogenic Yet Neglected Human Retrovirus: From Transmission to Therapeutic Treatment.
Viruses, 10(1):E1, 21 Dec 2017
Cited by: 53 articles | PMID: 29267225 | PMCID: PMC5795414
Review Free full text in Europe PMC
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Pseudotyping of HIV-1 with Human T-Lymphotropic Virus 1 (HTLV-1) Envelope Glycoprotein during HIV-1-HTLV-1 Coinfection Facilitates Direct HIV-1 Infection of Female Genital Epithelial Cells: Implications for Sexual Transmission of HIV-1.
mSphere, 3(2):e00038-18, 04 Apr 2018
Cited by: 4 articles | PMID: 29624497 | PMCID: PMC5885023
Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range.
J Virol, 65(1):162-169, 01 Jan 1991
Cited by: 195 articles | PMID: 1845882 | PMCID: PMC240501
Epstein-Barr virus permissively infects human syncytiotrophoblasts in vitro and induces replication of human T cell leukemia-lymphoma virus type I in dually infected cells.
Virology, 229(2):400-414, 01 Mar 1997
Cited by: 11 articles | PMID: 9126252
Cell-Free versus Cell-to-Cell Infection by Human Immunodeficiency Virus Type 1 and Human T-Lymphotropic Virus Type 1: Exploring the Link among Viral Source, Viral Trafficking, and Viral Replication.
J Virol, 90(17):7607-7617, 12 Aug 2016
Cited by: 30 articles | PMID: 27334587 | PMCID: PMC4988172
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: Z99 AI999999
NIAID NIH HHS (1)
Grant ID: Y99 AI999999



