Abstract
Free full text

A Bub1–Mad1 interaction targets the Mad1–Mad2 complex to unattached kinetochores to initiate the spindle checkpoint
Associated Data
Abstract
Recruitment of Mad1–Mad2 complexes to unattached kinetochores is a central event in spindle checkpoint signaling. Despite its importance, the mechanism that recruits Mad1–Mad2 to kinetochores is unclear. In this paper, we show that MAD-1 interacts with BUB-1 in Caenorhabditis elegans. Mutagenesis identified specific residues in a segment of the MAD-1 coiled coil that mediate the BUB-1 interaction. In addition to unattached kinetochores, MAD-1 localized between separating meiotic chromosomes and to the nuclear periphery. Mutations in the MAD-1 coiled coil that selectively disrupt interaction with BUB-1 eliminated MAD-1 localization to unattached kinetochores and between meiotic chromosomes, both of which require BUB-1, and abrogated checkpoint signaling. The identified MAD-1 coiled-coil segment interacted with a C-terminal region of BUB-1 that contains its kinase domain, and mutations in this region prevented MAD-1 kinetochore targeting independently of kinase activity. These results delineate an interaction between BUB-1 and MAD-1 that targets MAD-1–MAD-2 complexes to kinetochores and is essential for spindle checkpoint signaling.
Introduction
The spindle checkpoint generates a “wait anaphase” signal that inhibits the anaphase-promoting complex/cyclosome (APC/C; Musacchio and Salmon, 2007; Lara-Gonzalez et al., 2012). A key pathway component is the Mad1–Mad2 complex (Li and Murray, 1991; Hardwick and Murray, 1995), which concentrates at unattached kinetochores (Chen et al., 1998). Kinetochore-bound Mad1–Mad2 catalyzes a conformational transition of free Mad2 (Fig. 1 A; Mapelli and Musacchio, 2007; Luo and Yu, 2008) that promotes its association with the APC/C activator Cdc20 to restrict APC/C activity until microtubule attachment removes Mad1–Mad2 from the kinetochore.
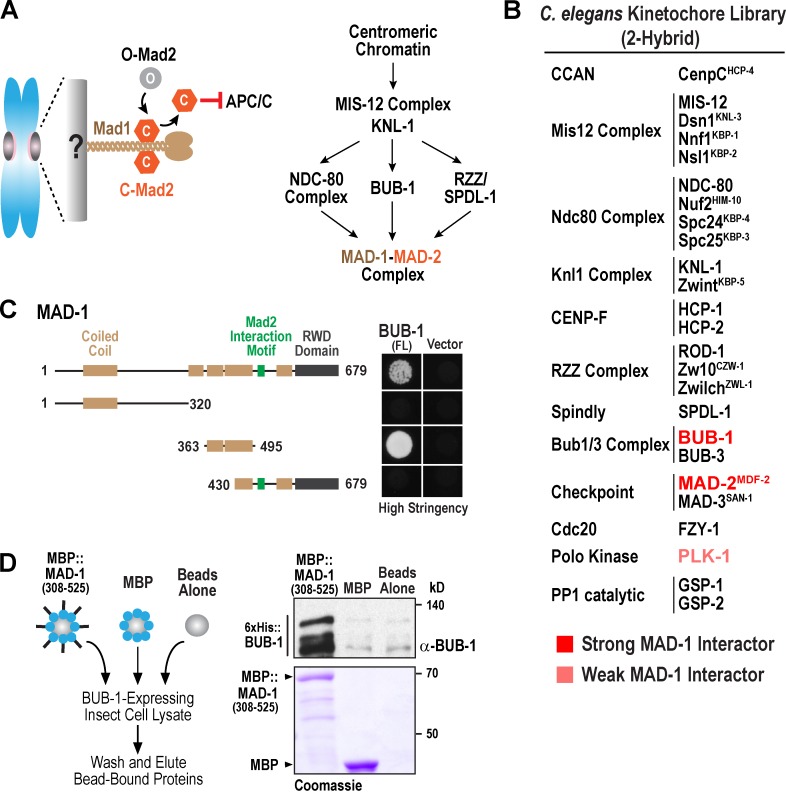
A kinetochore protein two-hybrid library screen identifies a MAD-1–BUB-1 interaction. (A, left) Schematic of checkpoint signaling mediated by kinetochore-anchored Mad1–Mad2 complexes. The inactive (O, open) and active (C, closed) conformers of Mad2 are depicted. (right) Summary of requirements for MAD-1–MAD-2 targeting to unattached kinetochores in C. elegans. (B) Summary of kinetochore protein two-hybrid screen. Strong indicates growth in low and high stringency conditions; weak indicates growth only under low stringency conditions. (C) Mapping of the region of MAD-1 that interacts with BUB-1 (FL). (D) Biochemical analysis of the MAD-1–BUB-1 interaction. Experimental schematic is on the left. Bead eluates were analyzed by anti–BUB-1 immunoblotting (top right) and Coomassie staining (bottom right). The data shown are representative of three experiments.
Despite progress in understanding checkpoint regulation of the APC/C, how Mad1–Mad2 targets to unattached kinetochores remains unclear. Mad1 is a coiled-coil protein with a Mad2 interaction motif (Chen et al., 1999) and a C-terminal RWD (RING finger, WD repeat, and DEAD-like proteins) domain, which is also found in other kinetochore proteins (Kim et al., 2012). The Mad1 RWD domain has been suggested to contribute to kinetochore targeting (Kim et al., 2012), but conclusive evidence is lacking. In vivo studies have shown that the conserved Bub1 kinase (Hoyt et al., 1991; Roberts et al., 1994; Taylor and McKeon, 1997) is required for Mad1–Mad2 kinetochore recruitment (Sharp-Baker and Chen, 2001; Gillett et al., 2004; Johnson et al., 2004). In budding yeast, Bub1 and Mad1 coimmunoprecipitate in the checkpoint-active state, and a region adjacent to the Bub1 kinase domain is required for checkpoint signaling (Brady and Hardwick, 2000; Warren et al., 2002). Mutations compromising Bub1 kinase activity reduce Mad1 kinetochore localization and spindle checkpoint signaling in human cells, but a Bub1 mutant lacking the kinase domain rescues checkpoint signaling in yeast and in Bub1-deleted mouse cells (Warren et al., 2002; Kang et al., 2008; Klebig et al., 2009; Perera and Taylor, 2010; Ricke et al., 2012). The microtubule-binding Ndc80 complex and the RZZ (Rod–Zw10–Zwilch) complex—which is conserved in metazoans but absent in fungi and plants—are also required for Mad1–Mad2 recruitment (Martin-Lluesma et al., 2002; Buffin et al., 2005; Kops et al., 2005). However, connections between any of these components and Mad1 have not been reported, leaving open the question of how they contribute to Mad1–Mad2 localization.
Here, we address the mechanisms that target Mad1–Mad2 to kinetochores in the Caenorhabditis elegans embryo. We identify an interaction between MAD-1 and BUB-1 in a two-hybrid screen of a kinetochore protein library. Analysis of precisely engineered mutations revealed that the BUB-1 interaction is required for MAD-1 kinetochore localization and checkpoint signaling. These results elucidate a direct recruitment mechanism for the Mad1–Mad2 complex to unattached kinetochores.
Results and discussion
A two-hybrid screen identifies BUB-1 as a MAD-1 interactor
Prior analysis of C. elegans MAD-1 and MAD-2 (also called MDF-1 and MDF-2; Kitagawa and Rose, 1999) revealed that MAD-1–MAD-2 kinetochore targeting requires the Ndc80 complex, the RZZ complex, and BUB-1 (Fig. 1 A; Gassmann et al., 2008; Yamamoto et al., 2008; Essex et al., 2009). The dynein recruitment factor SPDL-1 (Spindly) is also required in C. elegans, whereas it is not required in human and Drosophila melanogaster cells (Griffis et al., 2007; Gassmann et al., 2008, 2010; Yamamoto et al., 2008). To begin to understand this multifactorial recruitment reaction and determine whether it requires direct protein–protein interactions between MAD-1 and other kinetochore components, we performed a yeast two-hybrid screen with a library of kinetochore components. In addition to the expected interaction with MAD-2 (Fig. 1 B), MAD-1 exhibited a robust two-hybrid interaction with BUB-1 and a weaker interaction with the Polo kinase PLK-1 (Fig. 1 B). Most tested components exhibited at least one positive interaction (Fig. S1 A), reducing the likelihood that the absence of a MAD-1 interaction is a false negative. Given the conserved requirement for Bub1 in Mad1–Mad2 kinetochore recruitment, we focused on the MAD-1–BUB-1 interaction.
Additional two-hybrid analysis mapped the BUB-1 interaction to a segment of the MAD-1 coiled coil (aa 365–495; Fig. 1 C). Incubation of beads coated with purified maltose-binding protein (MBP)–MAD-1308–525 in insect cell lysate containing BUB-1 revealed an association of MBP–MAD-1308–525, but not MBP, with full-length (FL) and truncated BUB-1 (Fig. 1 D), validating the two-hybrid analysis. We were unable to detect an interaction when both components were purified, the reason for which is currently unclear (unpublished data). Thus, two-hybrid and biochemical analysis suggest that a segment of the MAD-1 coiled coil interacts with BUB-1.
MAD-1 localization to unattached kinetochores requires BUB-1 and repetitive segments in the KNL-1 N terminus
In C. elegans, MAD-1 is not detectable at kinetochores during unperturbed bipolar mitosis (Fig. S1 C) but is enriched at unattached kinetochores of monopolar spindles generated by depletion of the centriole duplication kinase ZYG-1 (Fig. 2 A; Essex et al., 2009). BUB-1 depletion reduces kinetochore MAD-1 to background levels (Fig. 2 A, Fig. S1 D, and Video 1). BUB-1 is recruited to kinetochores by the KNL-1 N terminus (Fig. 2 B; London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012), which contains a series of eight MELT repeats (Desai et al., 2003; Cheeseman et al., 2004; Vleugel et al., 2012). To determine whether BUB-1 must be present at kinetochores to recruit MAD-1, we used an RNAi-resistant KNL-1 transgenic system (Espeut et al., 2012) to engineer eight strains expressing wild-type (WT) or mutant forms of KNL-1 that delete or mutate different segments of the N terminus (Fig. 2 B and Fig. S1 B). We introduced a bub-1::gfp transgene into these strains and measured BUB-1::GFP relative to KNL-1::mCherry (mCh) on kinetochores of aligned chromosomes after endogenous KNL-1 depletion. The results revealed that progressive deletions of nonoverlapping regions of the KNL-1 N terminus had a graded effect on BUB-1 kinetochore recruitment (Fig. 2 B), with removal of a large N-terminal segment being necessary to abolish BUB-1 kinetochore localization and cause embryonic lethality (Δ85–505 and Δ136–505; Fig. 2 B). Thus, the N-terminal MELT repeats of C. elegans KNL-1 contribute additively to BUB-1 targeting, consistent with recent structural work (Primorac et al., 2013; Krenn et al., 2014) and studies in human cells (Vleugel et al., 2013; Zhang et al., 2013). A comparison of MAD-1 and BUB-1 recruitment in two KNL-1 mutants (Δ241–505 and Δ85–505) additionally revealed that MAD-1 localization to unattached kinetochores parallels that of BUB-1. Compared with WT KNL-1, the Δ241–505 KNL-1 mutant recruited comparably lower levels of MAD-1 and BUB-1, whereas the Δ85–505 KNL-1 mutant failed to recruit BUB-1 and MAD-1 (Fig. 2, C and D; and Videos 2 and 3). Thus, reducing BUB-1 localization at kinetochores by modifying KNL-1 leads to comparable reduction in MAD-1, consistent with a potential direct role for BUB-1 in MAD-1 kinetochore recruitment.
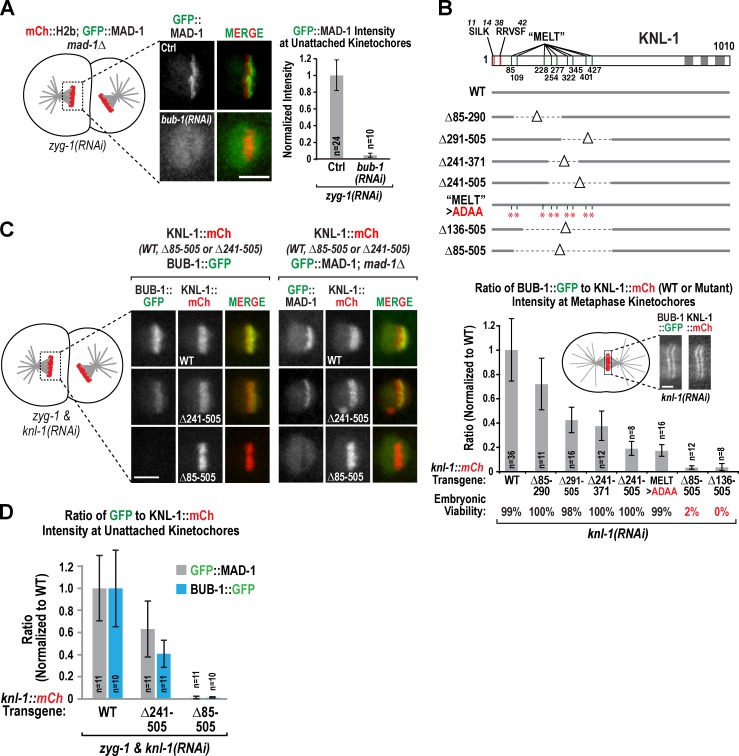
MAD-1 accumulation requires kinetochore-localized BUB-1. (A) The fluorescence intensity of GFP::MAD-1 at unattached kinetochores in monopolar spindles was quantified and expressed as a fraction of that in controls. n is the number of embryos; error bars are the 95% confidence interval of the mean. Ctrl, control. (B) Schematic showing the deleted/mutated regions in the indicated KNL-1 mutants. Graph plots the ratio of BUB-1::GFP to KNL-1::mCh for each KNL-1 variant normalized by dividing by the same ratio for WT KNL-1::mCh. Endogenous KNL-1 was depleted in all conditions. n is number of embryos analyzed. Error bars are the 95% confidence interval of the mean. See also Fig. S1 B. (C and D) MAD-1 and BUB-1 targeting to unattached kinetochores in strains expressing the indicated KNL-1 variants after depletion of ZYG-1 and endogenous KNL-1. n is the number of embryos analyzed. Graph plots the ratio of GFP::MAD-1 (in a mad-1Δ background) or BUB-1::GFP to KNL-1::mCh at kinetochores of monopolar spindles in the indicated strains. Error bars are the 95% confidence interval of the mean. Bars: (A and C) 5 µm; (B) 2 µm.
Identification of mutations in MAD-1 that selectively disrupt the MAD-1–BUB-1 interaction
To assess the functional significance of the MAD-1–BUB-1 interaction (Fig. 1, C and D), we performed unbiased mutagenesis of a MAD-1 fragment (aa 308–525) that includes the MAD-2 interaction motif (Fig. 3 A). This analysis identified 21 clones containing single amino acid changes in MAD-1308–525 that disrupt the MAD-1–BUB-1 two-hybrid interaction. These clones were rescreened versus BUB-1, MAD-2, and MAD-1308–525 (to assess self-interaction). The MAD-1 mutants disrupting interaction with BUB-1, but not MAD-2, clustered in a 74–amino acid segment of the coiled coil (Fig. 3 A and Fig. S2 A). However, the majority of these mutations also compromised MAD-1 self-interaction, potentially by disrupting coiled-coil structure. To overcome the effect of random mutations on coiled-coil structure, we took a directed approach by mutating the b, c, and f residues that extend away from the interior of the coiled coil to alanine (Fig. 3 B and Fig. S2 B; Lettman et al., 2013). Mutations in three heptads bracketing a predicted break in the coiled coil eliminated the BUB-1 interaction, and mutations in a fourth heptad compromised the interaction at high but not low stringency. In contrast, mutations in the other heptads or in the predicted break had no effect. We focused on heptad 6, for which we generated both 3A (b, c, and f residues) and D423A (f residue only) mutants that compromised the BUB-1 but not the MAD-2 or the MAD-1 self-interaction (Fig. 3 B and Fig. S2, C and D). The 3A mutation also eliminated the ability of MBP–MAD-1308–525–coated beads to pull down BUB-1 from insect cell lysate (Fig. 3 C). This region of the MAD-1 coiled coil is moderately conserved (Fig. S2 E), but functional analysis will be necessary to assess whether the interaction is conserved in other species.
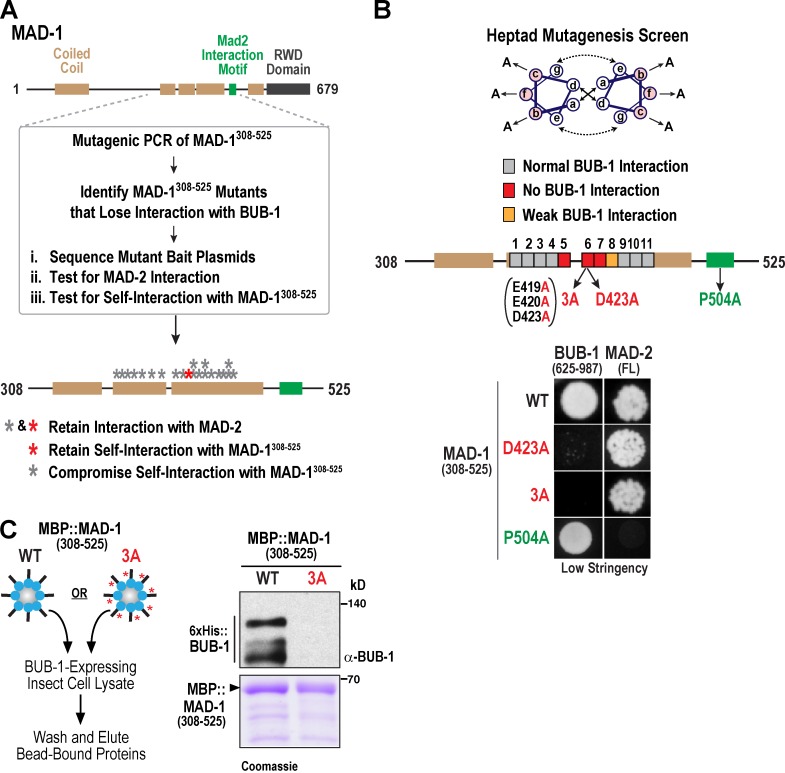
Unbiased and targeted mutagenesis of the MAD-1 coiled coil. (A) Schematic summarizing identification of MAD-1 mutants generated by unbiased mutagenesis that fail to bind BUB-1. See also Fig. S2 A. (B) Schematic of targeted heptad mutagenesis and summary of results. Mutations that resulted in no or weak BUB-1 binding are indicated. The P504A mutation prevents interaction with MAD-2. (C) Biochemical analysis of MAD-13A conducted as in Fig. 1 D. The blot and gel image is representative of three experiments.
In addition to specifically disrupting the interaction of MAD-1 with BUB-1, we used structural data (Sironi et al., 2002) to engineer a mutation that eliminates the interaction of MAD-1 with MAD-2 (Fig. 3 B, P504A). This mutation served as a control in functional assays, as it is expected to eliminate checkpoint signaling without affecting the MAD-1–BUB-1 interaction (Fig. 3 B).
MAD-1 mutants defective in interacting with BUB-1 do not localize to unattached kinetochores and fail to support checkpoint signaling
To assess the effect of MAD-1 mutants defective in interacting with BUB-1 in vivo, we generated single-copy gfp::mad-1 transgenes (Fig. S2 F) and introduced them into the mdf-1(gk2)–null mutant (referred to as mad-1Δ; Kitagawa and Rose, 1999). Immunoblotting confirmed replacement of endogenous MAD-1 with comparably expressed transgene-encoded GFP::MAD-1 variants (Fig. 4 A); all transgenes rescued mad-1Δ inviability and were propagated in the mad-1Δ background. We next analyzed MAD-1 localization on monopolar spindles (Fig. 4 B and Video 4). MAD-1P504A, which is defective in interacting with MAD-2, enriched at unattached kinetochores but, as it is checkpoint defective, did not delay the cell cycle; consequently, the extent of MAD-1P504A enrichment was lower than for MAD-1WT (Fig. 4, B and C). In comparison, MAD-13A and MAD-1D423A, which are defective in interacting with BUB-1, accumulated at kinetochores to a significantly reduced extent (Fig. 4, B and C). Thus, mutations that selectively disrupt interaction with BUB-1 reduce MAD-1 accumulation at kinetochores.
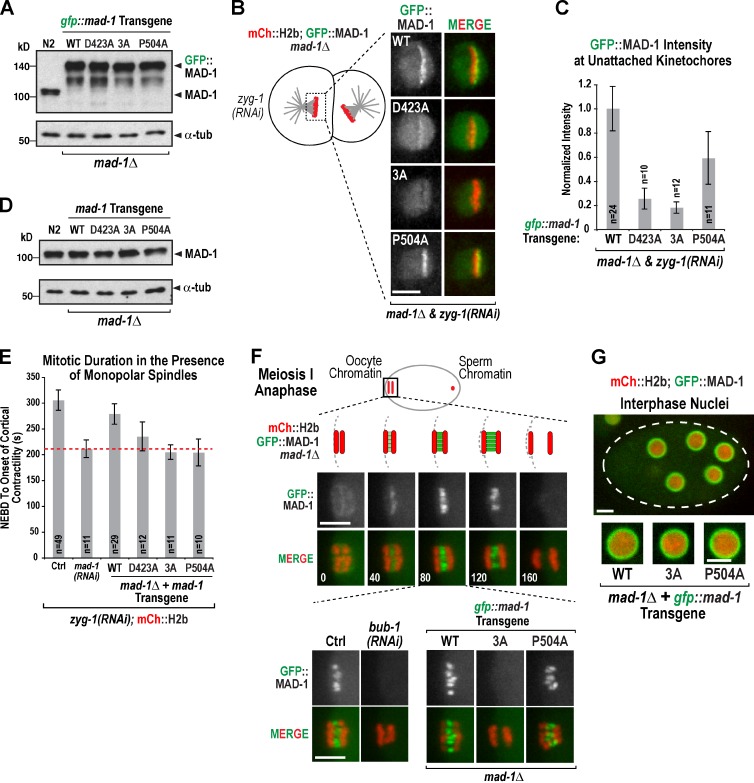
Analysis of BUB-1 interaction-defective MAD-1 mutants in vivo. (A) GFP::MAD-1 variants expressed from single-copy transgene insertions (Fig. S2 F), propagated in a mad-1Δ background, and analyzed by anti–MAD-1 immunoblotting. α-Tubulin (α-tub) serves as a loading control. (B and C) Images and quantification of GFP::MAD-1 variant localization to unattached kinetochores. Unpaired t tests show that MAD-13A is significantly reduced relative to MAD-1WT (P < 0.0001) and MAD-1P504 (P = 0.0004); MAD-1D423A is also significantly reduced (P < 0.0001 relative to MAD-1WT, and P = 0.0066 relative to MAD-1P504). The WT value is reproduced from Fig. 2 A. n is number of embryos analyzed. Error bars are the 95% confidence interval of the mean. (D) Immunoblotting of untagged MAD-1 variants. α-Tubulin serves as a loading control. (E) Quantification of time from NEBD to onset of cortical contractility in the presence of monopolar spindles. n is number of embryos analyzed. The red dotted line indicates mitotic duration in mad-1(RNAi), where the checkpoint is inactive. Error bars are the 95% confidence interval of the mean. (F) MAD-1 accumulation between separating homologous chromosomes in early anaphase of oocyte meiosis I. Time (seconds) on the bottom left of merge images are relative to anaphase onset. Images on the bottom left show that this localization depends on BUB-1. Images on the bottom right show loss of this localization for MAD-13A but not MAD-1P504A. 3–11 embryos were imaged per condition. Ctrl, control. (G) Nuclear periphery enrichment of MAD-1 for the indicated conditions. Dotted line indicates the embryo outline. More than eight embryos were imaged per condition. Bars, 5 µm.
Next, we measured the monopolar spindle-induced cell cycle delay (Essex et al., 2009) in the presence of the engineered MAD-1 mutants. As initial experiments revealed that GFP::MAD-1WT was compromised in its ability to induce a robust cell cycle delay (unpublished data), we reengineered MAD-1WT and all three mutants (3A, D423A, and P504A) as untagged transgenes. We verified expression of transgenes in a background containing mad-1Δ (Fig. 4 D), introduced an mCh::H2b marker, and measured the time from nuclear envelope breakdown (NEBD) to onset of cortical contractility in monopolar second divisions (Fig. 4 E). MAD-13A eliminated the cell cycle delay to the same degree as MAD-1 depletion (P = 0.5 compared with mad-1(RNAi), and P = 0.0001 compared with MAD-1WT in unpaired t tests), indicating loss of checkpoint signaling (Fig. 4 E). As expected, MAD-1P504A also abrogated the delay, whereas MAD-1D423A, which weakens kinetochore localization, had an intermediate effect (Fig. 4 E). Thus, the mutations in the MAD-1 coiled coil that selectively disrupt interaction with BUB-1 and perturb MAD-1 targeting to unattached kinetochores compromise checkpoint signaling.
BUB-1 interaction-defective MAD-1 fails to localize between separating chromosomes during anaphase of meiosis I
While imaging the GFP::MAD-1 strain, we noticed enrichment of MAD-1 between separating homologous chromosomes during anaphase of oocyte meiosis I (Fig. 4 F), a localization similar to that reported for BUB-1 in fixed analysis (Dumont et al., 2010). Simultaneous imaging revealed initial localization of BUB-1::mCh followed by GFP::MAD-1 at this site (Fig. S2 G). MAD-1 meiosis I anaphase localization required BUB-1 but not the NDC-80 or RZZ complexes, which are required for MAD-1 to localize to mitotic kinetochores (Fig. 4 F and Fig. S2 H). Similar to our analysis of unattached kinetochores, MAD-13A exhibited no localization in meiosis, whereas MAD-1P504A localized similarly to controls (Fig. 4 F). Although the functional significance of this nonkinetochore MAD-1 localization is currently unclear (Fig. S2 I), it supports the conclusion that the 3A mutant selectively disrupts the MAD-1–BUB-1 interaction rather than the currently poorly understood contributions of NDC-80 and RZZ to MAD-1 kinetochore targeting. MAD-1 also localized to the periphery of interphase nuclei (Fig. 4 G; Chen et al., 1998; Campbell et al., 2001; Iouk et al., 2002). This localization was independent of BUB-1, NDC-80, and RZZ (Fig. S2 J) and was unaffected by the MAD-13A and MAD-1P504A mutations (Fig. 4 G). Thus, both BUB-1–dependent localizations of MAD-1 in vivo, to unattached kinetochores and during anaphase of meiosis I, are compromised by the MAD-13A mutant, supporting the conclusion that the mutated region of MAD-1 mediates an interaction with BUB-1.
Alterations in the BUB-1 kinase domain perturb MAD-1 localization to unattached kinetochores independently of kinase activity
We next focused on the BUB-1 side of the MAD-1–BUB-1 interaction by narrowing the interaction domain to a C-terminal region of BUB-1 that includes its kinase domain (Fig. 5 A). We then used the single amino acid mutant MAD-1D423A (Fig. 3 B) to identify compensatory mutations in the BUB-1 C terminus that restored interaction by yeast two hybrid (Fig. 5 A). This effort identified a single mutation, N781D, which restored the interaction of BUB-1 with MAD-1D423A only under low stringency conditions (Fig. 5 A). No restoration was observed with MAD-13A (unpublished data). The conserved N781 residue of BUB-1 is located in a helix bracketing the active site with its side chain pointing toward the solvent (Fig. 5 B; Kang et al., 2008). In the adjacent turn of the helix is a conserved hydrophobic residue (L777) with its side chain also facing outward. N781D interacted with MAD-1, but the opposite charge substitution to N781K as well as an L777K mutation eliminated the interaction (Fig. 5 B).
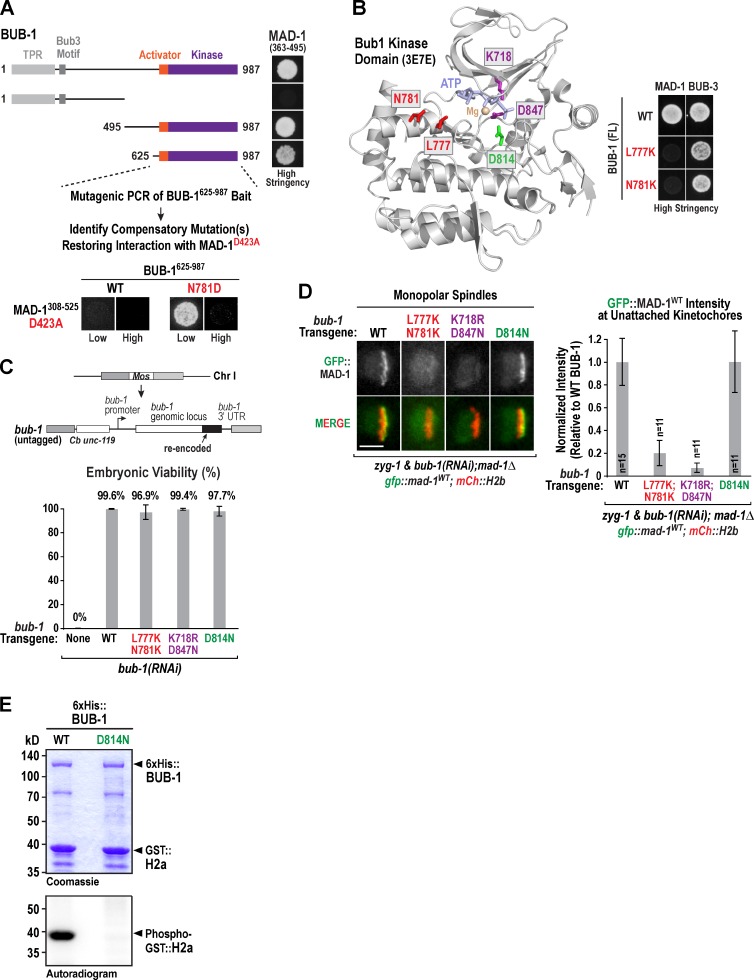
Identification and analysis of BUB-1 mutations affecting MAD-1 kinetochore7 localization. (A) Identification of the C-terminal region of BUB-1 as the interaction domain for MAD-1 and results of a compensatory two-hybrid screen using MAD-1D423A. TPR, tetratricopeptide repeat. (B) Location of N781 and other activity-related residues in the BUB-1 kinase domain (Protein Data Bank accession no. 3E7E; Kang et al., 2008). Residue numbering: Ce N781 (Hs N879), Ce L777 (Hs L875), Ce K718 (Hs K821), Ce K847 (Hs K946), and Ce D814 (Hs D917). Panel on the right shows two-hybrid analysis of the MAD-1–BUB-1 interaction with N781K and L777K mutations. (C) Schematic of bub-1 transgene and analysis of embryo viability after endogenous BUB-1 depletion for the indicated variants. At least 7 worms and >445 total embryos were scored per condition. Error bars are the 95% confidence interval of the mean. Chr I, chromosome I. (D) Images and quantification of GFP::MAD-1 at unattached kinetochores for the indicated BUB-1 variants, normalized relative to the WT transgene control. n is number of embryos analyzed; error bars are the 95% confidence interval of the mean. Bar, 5 µm. (E) Kinase activity assay of WT and D814N mutant BUB-1 with GST-fused C. elegans histone H2a as substrate. The blot and gel image are representative of three experiments.
To analyze the BUB-1 point mutants that perturbed interaction with MAD-1, we generated an RNAi-resistant bub-1 transgenic system (Fig. 5 C and Fig. S3 A) and used it to engineer a double L777K;N781K mutation (Fig. 5 C and Fig. S3B). As the helix containing L777K and N781K may participate in substrate binding, we also generated two additional mutant forms of BUB-1 perturbing kinase activity: K718R;D847N, which inhibits ATP and magnesium binding, and D814N, which mutates the catalytic aspartate residue of the HxD motif (Kang et al., 2008). Similar to BUB-1WT, all three mutant transgenes rescued embryonic lethality after BUB-1 depletion (Fig. 5 C). In addition, mCh-tagged versions of the K718R;D847N and D814N BUB-1 mutants integrated on chromosome II localized normally to kinetochores (unpublished data).
In contrast to BUB-1WT, the L777K;N781K mutant significantly compromised checkpoint signaling and MAD-1 accumulation at unattached kinetochores (Fig. 5 D, Fig. S3 C, and Video 5). Surprisingly, the two mechanism-based kinase mutants had strikingly different effects. The K718R;D847N mutant abolished MAD-1 kinetochore localization and abrogated checkpoint signaling; in contrast, the D814N mutant supported normal MAD-1 recruitment and checkpoint signaling (Fig. 5 D, Fig. S3 C, and Video 5). We confirmed that the D814N mutation abolished BUB-1 kinase activity, using GST-fused C. elegans histone H2a as a substrate (Fig. 5 E). Thus, the kinase activity of BUB-1 is not required for MAD-1 recruitment; however, the striking effect of the K718R;D847N mutation suggests that destabilization of the kinase domain may compromise interaction with MAD-1. A difference between kinase-dead Bub1 mutants has also been observed in mammals—a K796R (equivalent to C. elegans K718R) Bub1 mutant does not rescue checkpoint signaling in Bub1 mouse knockout cells, whereas a D920N (equivalent to C. elegans D847N) fully rescues (Taylor, S., personal communication; Perera and Taylor, 2010). In addition to serving as a cautionary note with respect to kinase-dead alleles, the C. elegans and mouse results suggest that alterations in the Bub1 kinase domain can influence Mad1 targeting independently of kinase activity, and this may in turn reflect a role for the conserved kinase domain of Bub1 in checkpoint regulation.
Conclusion
We have delineated and selectively perturbed a MAD-1–BUB-1 interaction that is important for kinetochore targeting of MAD-1 in C. elegans. This interaction is likely posttranslationally controlled, which would enable reversibility after microtubule attachment. A major question that remains to be addressed is how RZZ/SPDL-1 (Spindly) and NDC-80 contribute to MAD-1 kinetochore recruitment. The BUB-1K718R;D847N kinase domain mutation that abolishes MAD-1 targeting does not affect kinetochore recruitment of SPDL-1 (Fig. S3 D). In addition, BUB-1 depletion, which removes MAD-1, does not affect NDC-80 localization, and the MAD-13A mutant does not perturb BUB-1 localization (Fig. S3, E and F). Thus, disruption of the MAD-1–BUB-1 interaction using point mutants in either MAD-1 or BUB-1 drastically reduces MAD-1 kinetochore localization without affecting kinetochore pools of NDC-80 and RZZ/SPDL-1, suggesting that these other components may control the MAD-1–BUB-1 interaction. Addressing the conservation of the MAD-1 coiled coil–BUB-1 interaction and resolving the nature of the contributions of NDC-80 and RZZ/SPDL-1 to MAD-1 targeting will be important future goals.
Materials and methods
C. elegans strains
C. elegans strains (genotypes in Table S1) were maintained at 20°C. Engineered transgenes were cloned into pCFJ151 (GFP::MAD-1, MAD-1, BUB-1, and BUB-1::mCh) or pCFJ352 (BUB-1) and injected into strain EG6429 (MAD-1 transgenes), EG4322 (BUB-1, BUB-1::mCh, and KNL-1::mCh transgenes), or EG6701 (BUB-1 transgenes; Frøkjær-Jensen et al., 2008; Espeut et al., 2012). The amplified mad-1 genomic locus was flanked on the 5′ end by 5′-GATCGAATGAGACACGAAAC-3′ and on the 3′ end by 5′-ATAAGAAAATATATTTTCAG-3′. The amplified bub-1 genomic locus was flanked on the 5′ end by 5′-CTGAAATTAAGACTGGTTTA-3′ and on the 3′ end by 5′-CCAGCCATCCTCTTGTAAAA-3′. For the GFP::MAD-1 transgenes, the GFP sequence was inserted after the start codon and preceded by a CPGGGGGGT linker. For Bub1::mCh, the mCh was introduced before the stop codon after a TSVNGGRAGS linker. The sequence altered in BUB-1 to allow RNAi-mediated depletion of endogenous BUB-1 is shown in Fig. S3 A. For KNL-1::mCh, exon 4 was reencoded, and the mCh was introduced before the stop codon (Espeut et al., 2012). Single-copy insertion was confirmed by PCR. Transgenic strains were crossed into various marker or deletion strains using standard genetic procedures.
RNAi
Double-stranded RNAs (dsRNAs) were generated using oligonucleotides (oligos; Table S2) to amplify regions from N2 genomic DNA or cDNA. PCR reactions were used as templates for in vitro RNA production (Ambion), and the RNA was purified using a MEGAclear kit (Ambion). Eluted RNA from the T3 and T7 reactions were mixed together (40 µl each), combined with 40 µl of 3× soaking buffer (32.7 mM Na2HPO4, 16.5 mM KH2PO4, 6.3 mM KCl, and 14.1 mM NH4Cl), and annealed (68°C for 10 min and 37°C for 30 min). dsRNA was injected into L3/L4 hermaphrodite worms 37–47 h before imaging. For double or triple depletions, dsRNAs were mixed in equal amounts (≥0.65 mg/ml for each RNA).
Immunoblotting and immunofluorescence
For immunoblotting, a mixed population of worms growing at 20°C on a nematode growth medium + OP50 agar plate were collected with M9 + 0.1% Triton X-100, pelleted, and washed. Worms were vortexed in a mix of 100 µl M9 + 0.1% Triton X-100, 50 µl of 4× sample buffer, and 100 µl of glass beads, boiled, and then vortexed and boiled again. Samples were run on an SDS-PAGE gel, transferred to nitrocellulose, probed with 1 µg/ml affinity-purified anti–MAD-1 (rabbit; antigen was MAD-1 [aa 430–679]::6×His) or anti–α-tubulin (mouse monoclonal DM1-α; Sigma-Aldrich), and detected using an HRP-conjugated secondary antibody (rabbit or mouse; GE Healthcare). Sample loading was normalized using α-tubulin. For antibody production, MAD-1 (aa 430–679)::6×His was expressed in Escherichia coli, purified, and injected into rabbits (Covance). Serum was affinity purified on a HiTrap N-hydroxysuccinimide column to which MAD-1 (aa 430–679)::6×His was covalently coupled.
For Immunofluorescence, subbing solution was prepared by heating 100 ml of water to 60°C, adding 0.4 g gelatin (G-1890; Sigma-Aldrich), cooling the solution to 40°C, and adding 0.04 g chromalum (243361; Sigma-Aldrich); 100 mg poly-l-lysine (P-1524; Sigma-Aldrich) was added before stirring for 2–3 h and sterile filtering. Slides were prepared by heating subbing solution to 50–60°C, immersing the slides, and drying them overnight for use the next day. 10–13 gravid adult hermaphrodites were dissected into 3 µl of 0.8× egg salts made fresh from an egg salt solution (118 mM NaCl, 40 mM KCl, 3.4 mM MgCl2, 3.4 CaCl2, and 5 mM Hepes, pH 7.4) on a slide coated with subbing solution, overlaid with a coverslip, and immersed in liquid nitrogen. Coverslips were quickly removed and slides were immersed in −20°C methanol for 10–15 min. After washing with PBS, the slides were dried with a Kimwipe (Kimberly-Clark) except for the area containing the embryos. The embryo-containing area was circled with a PAP pen, and the embryos were blocked with AbDil (PBS, 0.1% Triton X-100, 0.1% NaN3, and 2% BSA) and incubated with a mix of Cy3-labeled BUB-1 antibody (rabbit; antigen was GST::BUB-1 [aa 287–665]; Desai et al., 2003) and an unlabeled α-tubulin antibody (mouse monoclonal DM1-α; Sigma-Aldrich), washed with PBS, and incubated with anti–mouse Cy5-labeled secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). After washing with PBS, antifade reagent with DAPI (ProLong gold; Invitrogen) was added, overlaid with a coverslip, and sealed with nail polish.
Yeast two-hybrid and mutagenic yeast two-hybrid screens
Yeast two-hybrid analysis was performed according to the manufacturer’s guidelines (Matchmaker; Takara Bio Inc.) High stringency refers to selection on −Leu/−Trp/−His/−Ade plates; low stringency refers to selection on −Leu/−Trp−His plates. Genes of interest were cloned from WT (N2) C. elegans cDNA and inserted into either pGADT7 or pGBKT7. Kinetochore screens were performed with MAD-1 (FL), (aa 1–320), (aa 308–495), and (aa 430–679) in pGADT7 and MAD-1 (aa 1–320), (aa 1–495), (aa 308–495), and (aa 430–679) in pGBKT7.
Mutagenic PCR was performed on MAD-1 (aa 308–525) in pGADT7 using oligos 5′-TAATACGACTCACTATAGGGC-3′ and 5′-AGATGGTGCACGATGCACAG-3′. 40 independent 25-µl PCR reactions (15 cycles) using Standard Taq Polymerase (New England Biolabs, Inc.) were combined and gel purified (Umezu et al., 1998). An equal amount of the pooled PCR library was mixed with restriction-digested MAD-1 (aa 308–525) pGADT7 plasmid, and transformed into yeast containing BUB-1 (aa 625–987) cloned in pGBKT7. Yeast were plated on −Leu/−Trp dropout plates, and then, replica were plated onto −Leu/−Trp and −Leu/−Trp/−Ade/−His plates. Colonies that failed to grow on the quadruple dropout plate were recovered from the −Leu/−Trp plates, and the plasmids were isolated and sequenced. MAD-1 mutants containing single amino acid changes were screened by yeast two hybrid for interaction with MAD-2 (FL) and MAD-1 (aa 308–525) cloned in pGBKT7. Mutants that maintained binding with MAD-1 (aa 308–525) pGBKT7 were screened against MAD-1 (aa 308–525) pGBKT7 harboring the identical mutation. For the compensatory screen, oligos used for PCR BUB-1 (aa 625–987) were 5′-CATCGGAAGAGAGTAGTAACAAAG-3′ and 5′-TTTTCGTTTTAAAACCTAAGAGTC-3′. A mix containing the pooled gel-purified PCR product library and cut plasmid (Bub1 [aa 625–987] pGBKT7) was transformed into yeast harboring MAD-1 D423A (aa 308–525) pGADT7. Plasmids were isolated from colonies that grew on selective media and sequenced.
Imaging and quantification
For all experiments, except the ones shown in Fig. 4 E and Fig. S3 F, images were acquired using a confocal imaging system (Revolution XD Confocal System; Andor Technology) with a confocal scanner unit (CSU-10; Yokogawa Corporation of America) mounted on an inverted microscope (TE2000-E; Nikon) equipped with solid-state 100-mW lasers, 60×, 1.4 NA Plan Apochromat and 100×, 1.4 NA Plan Apochromat lens, and an electron multiplication back-thinned charge-coupled device camera (binning 1 × 1; iXon; Andor Technology). Imaging for the experiment shown in Fig. 4 E was performed on an inverted system (Axio Observer.Z1; Carl Zeiss) with a spinning-disk confocal head (CSU-X1; Yokogawa Corporation of America), a 63×, 1.4 NA Plan Apochromat objective, and an electron-multiplying charge-coupled device camera (QuantEM: 512SC; Photometrics). Imaging for the experiment shown in Fig. S3 F was performed using a 100×, 1.35 NA U Plan Apochromat oil objective lens (Olympus) on a DeltaVision system (Applied Precision) built on a microscope (IX70; Olympus) and equipped with a charge-coupled device camera (CoolSNAP; Roper Scientific). 80 z sections at 0.2-µm steps were acquired and computationally deconvolved using softWoRx software (Applied Precision). Environmental temperatures during experimental acquisitions averaged 22°C for the Andor Technology system, 19°C for the Carl Zeiss system, and 21°C for the DeltaVision system.
For live imaging of one- and two-cell embryos, gravid hermaphrodite adult worms were dissected into M9 buffer, and embryos were manually transferred to 2% agarose pads and overlaid with a coverslip. To monitor unattached kinetochores (Fig. 2, A and C; Fig. 4 B; Fig. 5 D; and Fig. S3 D), a 6 × 2–µm z series was collected every 15 s in two-cell embryos. For nuclear pore localization (Fig. 4 G and Fig. S2 J), embryos were imaged using a 10 × 2–µm z series. For BUB-1::GFP/KNL-1::mCh (Fig. 2 B and Fig. S1 B) and Nuf2HIM-10::mCh (Fig. S3 E), one-cell embryos were imaged every 20 s using a 5 × 2–µm z series. For the monopolar spindle-induced mitotic delay measurements, two-cell embryos were imaged every 15 s (Fig. 4 E) or 20 s (Fig. S3 C) with a 5 × 2–µm z series.
For meiotic imaging experiments (Fig. 4 F and Fig. S2, G and H), gravid adult hermaphrodites were dissected into 0.8× egg salts made fresh from an egg salt solution (118 mM NaCl, 40 mM KCl, 3.4 mM MgCl2, 3.4 CaCl2, and 5 mM Hepes, pH 7.4), mounted in a microdevice designed for C. elegans embryos (Carvalho et al., 2011), and imaged at 100× every 20 s with a 5 × 2–µm z series. For mad-1Δ meiosis I imaging (Fig. S2 I), homozygous F1 mad-1Δ worms were picked from the progeny of a balanced mad-1Δ heterozygote, based on the absence of the GFP marker inserted on the balancer, and dissected to visualize oocyte meiosis I in the recently fertilized F2 mad-1Δ zygotes.
All images and videos were processed, scaled, and analyzed using ImageJ (National Institutes of Health), Photoshop (Adobe), and MetaMorph (Molecular Devices). To minimize bleaching of GFP::MAD-1 on monopolar spindles, imaging was initiated around the time MAD-1 forms a “cloud,” and maximum intensity projections of the z series were generated. The frame in which chromosomes aligned to form a pseudometaphase plate was identified and used to measure kinetochore intensity as described in Fig. S1 D; previous imaging of an mCh::H2b strain was used to confirm that the time of chromosome alignment was similar for the different mutants analyzed. For example, the time from NEBD to chromosome alignment (seconds) was 64 ± 9 for MAD-1WT, 66 ± 17 for MAD-1D423A, 57 ± 14 for MAD-13A, and 69 ± 14 for MAD-1P504A (error is 95% confidence interval of the mean; n > 10 for each). For Fig. 2 D, KNL-1::mCh at unattached kinetochores on monopolar spindles was measured on the same frame used for GFP::MAD-1 quantification. Quantification of BUB-1::GFP; KNL-1::mCh in Fig. 2 D was performed similarly.
Quantification of KNL-1, BUB-1, and Nuf2HIM-10 kinetochore localization during metaphase of one-cell embryos and SPDL-1 on monopolar two-cell embryos was performed on maximum intensity projections as schematized in Fig. S1 B. A rectangle was drawn around the kinetochore signal, and mean pixel intensity was measured. The rectangle was expanded by 5 pixels on all sides, and the difference in integrated intensity between the expanded rectangle and the original rectangle was used to define the background intensity per pixel (Fig. S1 B). Integrated kinetochore fluorescence was then calculated for the original rectangle after background subtraction.
Mitotic duration in the presence of monopolar spindles was measured as the interval between NEBD to onset of cortical contractility. NEBD was identified by the equilibration of the mCh::H2B signal between the nucleus and cytosol; onset of cortical contractility was identified as the first frame when persistent blebs formed on the cortex of the embryo.
Protein purification and binding analysis
MAD-1 (aa 308–525) and MAD-1 3A (aa 308–525) were cloned into pMal-c2X. MBP::6×His, H2A, and 6×His::BUB-1 were cloned into pET21a, pGEX6p-1, and pFastBac1, respectively. MAD-1 (aa 308–525) and MBP::6×His were expressed in BL21(DE3). MAD-1 3A (aa 308–525) and H2A were expressed in BL21(DE3)pLysS. E. coli cultures were grown to OD 0.6–0.8 and induced with 0.1 mM IPTG for 4 h at 20°C. 6×His::Bub1 pFastBac1 was transformed into DH10EMBacY. Baculovirus was generated following standard procedures (MultiBac baculovirus expression vector system [Epigenesys]; Bac-to-Bac [Invitrogen]). High Five cells were grown to 1–2 × 106 cells/ml and infected with V1 or V2 virus at 1:30 dilution for 48 h at 27°C.
Binding assays were performed by lysing E. coli and insect cells in 5 ml of 50 mM Tris, pH 7.6, 100 mM KCl, 1:100 lysozyme, 1:100 benzamidine, 10% glycerol, 5 mM 2-mercaptoethanol, 2 mM MgCl2, 0.2 mM ATP, 50 mM NaF, 1 mM Na3VO4, and a cOmplete EDTA-free protease inhibitor cocktail tablet (Roche). Lysates were pelleted, and a soluble fraction was bound to Amylose beads at 4°C for 30 min. Beads were washed, added to insect cell lysate containing 6×His::BUB-1, and rotated at 4°C for 1 h. Beads were washed and boiled in sample buffer. Bead-bound proteins were analyzed by Coomassie staining and immunoblotting using 1 µg/ml affinity-purified antibody to BUB-1 (rabbit; antigen was GST::BUB-1 [aa 287–665]; Desai et al., 2003).
Kinase assay
BUB-1 D814N mutant virus was generated as noted (see Protein purification and binding analysis). High Five cells were grown to 1–1.75 × 106 cells/ml and infected with virus for 48 h at 27°C. His and GST purifications were performed using standard procedures. 6×His::BUB-1 and GST::H2A were dialyzed into 50 mM Tris, pH 7.88, 150 mM KCl, 10% glycerol, 2 mM MgCl2, 5 mM 2-mercaptoethanol, and 0.2 mM ATP. Kinase reactions were performed with 0.35 µM BUB-1 and 3.5 µM H2A (1:10 ratio) with 12 µCi γ-[32P]ATP for 1 h at room temperature. After overnight exposure, the phosphorimager screen was viewed using a Personal Molecular Imager (Bio-Rad Laboratories).
Online supplemental material
Fig. S1 describes the yeast two-hybrid library, quantification of expression of KNL-1 variants, localization of MAD-1 during bipolar mitosis, and method used to quantify MAD-1 localization at unattached kinetochores. Fig. S2 shows hits from the mutagenic yeast two-hybrid screen, MAD-1 coiled-coil sequence features, two-hybrid analysis with FL proteins, confirmation that mutants tested do not affect MAD-1 self-interaction, analysis of MAD-1 localization and function during meiosis I, and MAD-1 nuclear periphery localization. Fig. S3 describes the BUB-1 transgene system, analysis of checkpoint signaling in BUB-1 mutants, and analysis of SPDL-1, NDC-80, and BUB-1 kinetochore localization. Tables S1 and S2 show C. elegans strains (Table S1) and oligos used to generate the dsRNAs (Table S2) used in this study. Video 1 shows loss of MAD-1 kinetochore localization after BUB-1 depletion. Videos 2 and 3 show localization of BUB-1 and MAD-1, respectively, in the indicated KNL-1 variants after endogenous KNL-1 depletion. Video 4 shows localization of engineered MAD-1 mutants in a mad-1Δ background. Video 5 shows localization of MAD-1 in the presence of WT and engineered BUB-1 mutants after depletion of endogenous BUB-1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201311015/DC1.
Acknowledgments
We thank S. Biggins and S. Taylor for sharing unpublished data, R. Kitagawa and M. Sarov/T. Hyman for strains, R. Kolodner for advice on mutagenic PCR, N. Pogliano and A. Gleason for technical assistance, and R. Green, C. Campbell, and P. Gonzalez for comments on the manuscript.
This work was supported by a National Institutes of Health grant (GM074215) to A. Desai. M. Moyle was supported by a Training in Cancer Cell Biology grant (National Institutes of Health–National Cancer Institute T32-CA067754) and a National Institutes of Health Predoctoral Genetics Training grant (T32 GM008666). A. Desai and K. Oegema receive salary and other support from the Ludwig Institute for Cancer Research.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- dsRNA
- double-stranded RNA
- FL
- full length
- MBP
- maltose-binding protein
- mCh
- mCherry
- NEBD
- nuclear envelope breakdown
- WT
- wild type
References
- Brady D.M., Hardwick K.G. 2000. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 10:675–678 10.1016/S0960-9822(00)00515-7 [Abstract] [CrossRef] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861 10.1016/j.cub.2005.03.052 [Abstract] [CrossRef] [Google Scholar]
- Campbell M.S., Chan G.K., Yen T.J. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953–963 [Abstract] [Google Scholar]
- Carvalho A., Olson S.K., Gutierrez E., Zhang K., Noble L.B., Zanin E., Desai A., Groisman A., Oegema K. 2011. Acute drug treatment in the early C. elegans embryo. PLoS ONE. 6:e24656 10.1371/journal.pone.0024656 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R., III, Oegema K., Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268 10.1101/gad.1234104 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., Murray A.W. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295 10.1083/jcb.143.2.283 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Chen R.H., Brady D.M., Smith D., Murray A.W., Hardwick K.G. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 10:2607–2618 10.1091/mbc.10.8.2607 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Dumont J., Oegema K., Desai A. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901 10.1038/ncb2093 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Espeut J., Cheerambathur D.K., Krenning L., Oegema K., Desai A. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482 10.1083/jcb.201111107 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Essex A., Dammermann A., Lewellyn L., Oegema K., Desai A. 2009. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 20:1252–1267 10.1091/mbc.E08-10-1047 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Frøkjær-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383 10.1038/ng.248 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R., III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399 10.1101/gad.1687508 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., Desai A. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971 10.1101/gad.1886810 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gillett E.S., Espelin C.W., Sorger P.K. 2004. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J. Cell Biol. 164:535–546 10.1083/jcb.200308100 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Griffis E.R., Stuurman N., Vale R.D. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015 10.1083/jcb.200702062 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hardwick K.G., Murray A.W. 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131:709–720 10.1083/jcb.131.3.709 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [Abstract] [CrossRef] [Google Scholar]
- Iouk T., Kerscher O., Scott R.J., Basrai M.A., Wozniak R.W. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807–819 10.1083/jcb.200205068 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589 10.1242/jcs.01006 [Abstract] [CrossRef] [Google Scholar]
- Kang J., Yang M., Li B., Qi W., Zhang C., Shokat K.M., Tomchick D.R., Machius M., Yu H. 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell. 32:394–405 10.1016/j.molcel.2008.09.017 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kim S., Sun H., Tomchick D.R., Yu H., Luo X. 2012. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl. Acad. Sci. USA. 109:6549–6554 10.1073/pnas.1118210109 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kitagawa R., Rose A.M. 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1:514–521 10.1038/70309 [Abstract] [CrossRef] [Google Scholar]
- Klebig C., Korinth D., Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858 10.1083/jcb.200902128 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R., III, Tagaya M., Cleveland D.W. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60 10.1083/jcb.200411118 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Krenn V., Overlack K., Primorac I., van Gerwen S., Musacchio A. 2014. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr. Biol. 24:29–39 10.1016/j.cub.2013.11.046 [Abstract] [CrossRef] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., Taylor S.S. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980 10.1016/j.cub.2012.10.006 [Abstract] [CrossRef] [Google Scholar]
- Lettman M.M., Wong Y.L., Viscardi V., Niessen S., Chen S.H., Shiau A.K., Zhou H., Desai A., Oegema K. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell. 25:284–298 10.1016/j.devcel.2013.03.011 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li R., Murray A.W. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531 10.1016/0092-8674(81)90015-5 [Abstract] [CrossRef] [Google Scholar]
- London N., Ceto S., Ranish J.A., Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906 10.1016/j.cub.2012.03.052 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Luo X., Yu H. 2008. Protein metamorphosis: the two-state behavior of Mad2. Structure. 16:1616–1625 10.1016/j.str.2008.10.002 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mapelli M., Musacchio A. 2007. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr. Opin. Struct. Biol. 17:716–725 10.1016/j.sbi.2007.08.011 [Abstract] [CrossRef] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., Nigg E.A. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270 10.1126/science.1075596 [Abstract] [CrossRef] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [Abstract] [CrossRef] [Google Scholar]
- Perera D., Taylor S.S. 2010. Sgo1 establishes the centromeric cohesion protection mechanism in G2 before subsequent Bub1-dependent recruitment in mitosis. J. Cell Sci. 123:653–659 10.1242/jcs.059501 [Abstract] [CrossRef] [Google Scholar]
- Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., Musacchio A. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2:e01030 10.7554/eLife.01030 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ricke R.M., Jeganathan K.B., Malureanu L., Harrison A.M., van Deursen J.M. 2012. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J. Cell Biol. 199:931–949 10.1083/jcb.201205115 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Roberts B.T., Farr K.A., Hoyt M.A. 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 14:8282–8291 [Europe PMC free article] [Abstract] [Google Scholar]
- Sharp-Baker H., Chen R.H. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250 10.1083/jcb.153.6.1239 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899 10.1016/j.cub.2012.03.051 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.T., Musacchio A. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506 10.1093/emboj/21.10.2496 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Taylor S.S., McKeon F. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 89:727–735 10.1016/S0092-8674(00)80255-X [Abstract] [CrossRef] [Google Scholar]
- Umezu K., Sugawara N., Chen C., Haber J.E., Kolodner R.D. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 148:989–1005 [Europe PMC free article] [Abstract] [Google Scholar]
- Vleugel M., Hoogendoorn E., Snel B., Kops G.J. 2012. Evolution and function of the mitotic checkpoint. Dev. Cell. 23:239–250 10.1016/j.devcel.2012.06.013 [Abstract] [CrossRef] [Google Scholar]
- Vleugel M., Tromer E., Omerzu M., Groenewold V., Nijenhuis W., Snel B., Kops G.J. 2013. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J. Cell Biol. 203:943–955 10.1083/jcb.201307016 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Warren C.D., Brady D.M., Johnston R.C., Hanna J.S., Hardwick K.G., Spencer F.A. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 13:3029–3041 10.1091/mbc.E02-04-0203 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yamagishi Y., Yang C.H., Tanno Y., Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752 10.1038/ncb2515 [Abstract] [CrossRef] [Google Scholar]
- Yamamoto T.G., Watanabe S., Essex A., Kitagawa R. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 183:187–194 10.1083/jcb.200805185 [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Zhang G., Lischetti T., Nilsson J. 2013. A minimal number of MELT repeats supports all functions of KNL1 in chromosome segregation. J. Cell Sci. In press 10.1242/jcs.139725 [Abstract] [CrossRef] [Google Scholar]
Articles from The Journal of Cell Biology are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1083/jcb.201311015
Read article for free, from open access legal sources, via Unpaywall:
http://jcb.rupress.org/content/jcb/204/5/647.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Centromeres in cancer: Unraveling the link between chromosomal instability and tumorigenesis.
Med Oncol, 41(11):254, 01 Oct 2024
Cited by: 0 articles | PMID: 39352464
Review
CCDC68 Maintains Mitotic Checkpoint Activation by Promoting CDC20 Integration into the MCC.
Adv Sci (Weinh), 11(35):e2406009, 17 Jul 2024
Cited by: 1 article | PMID: 39018254 | PMCID: PMC11425217
The outer kinetochore components KNL-1 and Ndc80 complex regulate axon and neuronal cell body positioning in the C. elegans nervous system.
Mol Biol Cell, 35(6):ar83, 24 Apr 2024
Cited by: 0 articles | PMID: 38656792 | PMCID: PMC11238089
<i>FBXO42</i> activity is required to prevent mitotic arrest, spindle assembly checkpoint activation and lethality in glioblastoma and other cancers.
NAR Cancer, 6(2):zcae021, 20 May 2024
Cited by: 0 articles | PMID: 38774470 | PMCID: PMC11106029
Coregulation of NDC80 Complex Subunits Determines the Fidelity of the Spindle-Assembly Checkpoint and Mitosis.
Mol Cancer Res, 22(5):423-439, 01 May 2024
Cited by: 2 articles | PMID: 38324016 | PMCID: PMC11063766
Go to all (78) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores.
EMBO Rep, 15(3):291-298, 29 Jan 2014
Cited by: 39 articles | PMID: 24477934 | PMCID: PMC3989695
TRIP13PCH-2 promotes Mad2 localization to unattached kinetochores in the spindle checkpoint response.
J Cell Biol, 211(3):503-516, 02 Nov 2015
Cited by: 28 articles | PMID: 26527744 | PMCID: PMC4639874
The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells.
Open Biol, 5(11):150160, 01 Nov 2015
Cited by: 33 articles | PMID: 26581576 | PMCID: PMC4680571
The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit.
Prog Cell Cycle Res, 5:431-439, 01 Jan 2003
Cited by: 37 articles | PMID: 14593737
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: T32-CA067754
Grant ID: T32 CA067754
NIGMS NIH HHS (3)
Grant ID: GM074215
Grant ID: R01 GM074215
Grant ID: T32 GM008666

 1,2
1,2



