Abstract
Unlabelled
It is generally acknowledged that human broadly neutralizing antibodies (bNAbs) capable of neutralizing multiple HIV-1 clades are often polyreactive or autoreactive. Whereas polyreactivity or autoreactivity has been proposed to be crucial for neutralization breadth, no systematic, quantitative study of self-reactivity among nonneutralizing HIV-1 Abs (nNAbs) has been performed to determine whether poly- or autoreactivity in bNAbs is a consequence of chronic antigen (Ag) exposure and/or inflammation or a fundamental property of neutralization. Here, we use protein microarrays to assess binding to >9,400 human proteins and find that as a class, bNAbs are significantly more poly- and autoreactive than nNAbs. The poly- and autoreactive property is therefore not due to the infection milieu but rather is associated with neutralization. Our observations are consistent with a role of heteroligation for HIV-1 neutralization and/or structural mimicry of host Ags by conserved HIV-1 neutralization sites. Although bNAbs are more mutated than nNAbs as a group, V(D)J mutation per se does not correlate with poly- and autoreactivity. Infrequent poly- or autoreactivity among nNAbs implies that their dominance in humoral responses is due to the absence of negative control by immune regulation. Interestingly, four of nine bNAbs specific for the HIV-1 CD4 binding site (CD4bs) (VRC01, VRC02, CH106, and CH103) bind human ubiquitin ligase E3A (UBE3A), and UBE3A protein competitively inhibits gp120 binding to the VRC01 bNAb. Among these four bNAbs, avidity for UBE3A was correlated with neutralization breadth. Identification of UBE3A as a self-antigen recognized by CD4bs bNAbs offers a mechanism for the rarity of this bNAb class.Importance
Eliciting bNAbs is key for HIV-1 vaccines; most Abs elicited by HIV-1 infection or immunization, however, are strain specific or nonneutralizing, and unsuited for protection. Here, we compare the specificities of bNAbs and nNAbs to demonstrate that bNAbs are significantly more poly- and autoreactive than nNAbs. The strong association of poly- and autoreactivity with bNAbs, but not nNAbs from infected patients, indicates that the infection milieu, chronic inflammation and Ag exposure, CD4 T-cell depletion, etc., alone does not cause poly- and autoreactivity. Instead, these properties are fundamentally linked to neutralization breadth, either by the requirement for heteroligation or the consequence of host mimicry by HIV-1. Indeed, we show that human UBE3A shares an epitope(s) with HIV-1 envelope recognized by four CD4bs bNAbs. The poly- and autoreactivity of bNAbs surely contribute to the rarity of membrane-proximal external region (MPER) and CD4bs bNAbs and identify a roadblock that must be overcome to induce protective vaccines.Free full text

Polyreactivity and Autoreactivity among HIV-1 Antibodies
ABSTRACT
It is generally acknowledged that human broadly neutralizing antibodies (bNAbs) capable of neutralizing multiple HIV-1 clades are often polyreactive or autoreactive. Whereas polyreactivity or autoreactivity has been proposed to be crucial for neutralization breadth, no systematic, quantitative study of self-reactivity among nonneutralizing HIV-1 Abs (nNAbs) has been performed to determine whether poly- or autoreactivity in bNAbs is a consequence of chronic antigen (Ag) exposure and/or inflammation or a fundamental property of neutralization. Here, we use protein microarrays to assess binding to >9,400 human proteins and find that as a class, bNAbs are significantly more poly- and autoreactive than nNAbs. The poly- and autoreactive property is therefore not due to the infection milieu but rather is associated with neutralization. Our observations are consistent with a role of heteroligation for HIV-1 neutralization and/or structural mimicry of host Ags by conserved HIV-1 neutralization sites. Although bNAbs are more mutated than nNAbs as a group, V(D)J mutation per se does not correlate with poly- and autoreactivity. Infrequent poly- or autoreactivity among nNAbs implies that their dominance in humoral responses is due to the absence of negative control by immune regulation. Interestingly, four of nine bNAbs specific for the HIV-1 CD4 binding site (CD4bs) (VRC01, VRC02, CH106, and CH103) bind human ubiquitin ligase E3A (UBE3A), and UBE3A protein competitively inhibits gp120 binding to the VRC01 bNAb. Among these four bNAbs, avidity for UBE3A was correlated with neutralization breadth. Identification of UBE3A as a self-antigen recognized by CD4bs bNAbs offers a mechanism for the rarity of this bNAb class.
IMPORTANCE Eliciting bNAbs is key for HIV-1 vaccines; most Abs elicited by HIV-1 infection or immunization, however, are strain specific or nonneutralizing, and unsuited for protection. Here, we compare the specificities of bNAbs and nNAbs to demonstrate that bNAbs are significantly more poly- and autoreactive than nNAbs. The strong association of poly- and autoreactivity with bNAbs, but not nNAbs from infected patients, indicates that the infection milieu, chronic inflammation and Ag exposure, CD4 T-cell depletion, etc., alone does not cause poly- and autoreactivity. Instead, these properties are fundamentally linked to neutralization breadth, either by the requirement for heteroligation or the consequence of host mimicry by HIV-1. Indeed, we show that human UBE3A shares an epitope(s) with HIV-1 envelope recognized by four CD4bs bNAbs. The poly- and autoreactivity of bNAbs surely contribute to the rarity of membrane-proximal external region (MPER) and CD4bs bNAbs and identify a roadblock that must be overcome to induce protective vaccines.
INTRODUCTION
A major obstacle in HIV-1 vaccine research is the inability to elicit broadly neutralizing antibodies (bNAbs) that recognize stable, neutralizing epitopes present on the envelope (Env) proteins of multiple viral clades (1). Indeed, neither vaccination nor active infection routinely elicits bNAbs, and high bNAb titers arise only in ~20% of infected individuals after 2 to 3 years of infection (2, 3). Though rare, many bNAbs have been recently isolated as a consequence of technological advances (4, 5). These bNAbs target one of four conserved sites on HIV-1 Env, including the gp120 CD4 binding site (CD4bs) (5,–7), gp41 membrane-proximal external region (MPER) (8,–10), gp120 V1/V2 loop (4, 11), and gp120 V3-glycans (12).
Many factors are hypothesized to restrict the development of bNAbs, including the rapid evolution of HIV-1 Env proteins (13), extensive Env glycosylation (14), conformational masking (15), and the scarcity of critical Env epitopes (16). Haynes et al. (17) first noted polyreactivity and autoreactivity among bNAbs and hypothesized that conserved, neutralizing epitopes of HIV-1 might mimic host proteins to avoid robust humoral responses by the removal/inactivation of responder B cells through immunological tolerance. Our characterization of two MPER-binding bNAbs, 2F5 and 4E10, identified two human proteins, kynureninase (KYNU) and splicing factor 3b subunit 3 (SF3B3), as autoantigens mimicked by the 2F5 and 4E10 HIV-1 epitopes (18). Mimicry of these conserved self-antigens by HIV-1 suggests that bNAbs to the 2F5 and 4E10 epitopes are proscribed by immune tolerance, a notion supported by impaired B-cell development in knock-in mice expressing the 2F5 or 4E10 VHDJH and VLJL rearrangements (19,–21). Further, immunization of opossums that naturally carry a KYNU mutation abrogating the shared 2F5 HIV-1 epitope elicited extraordinary Ab titers to the gp41 2F5 motif missing in opossums (18). It remains unclear whether other classes of HIV-1 bNAbs or nonneutralizing HIV-1 Abs (nNAbs) are also influenced by immunological tolerance.
In addition to specific autoreactivity, a number of bNAbs have been reported to be polyreactive, including the MPER bNAb 4E10 (18), and the CD4bs bNAbs CH103, CH106 (7), and CH98 (22). In contrast to autoreactive Abs that bind specific self-epitopes, polyreactive Abs are promiscuous binders of apparently dissimilar self- and nonself-antigens (23). During normal B-cell ontogeny, poly- and autoreactivity are common in early B-cell types, such as early immature, immature, and transitional B-cell compartments, but are significantly more rare in the mature B-cell compartment (24). Structural features strongly linked with Ab polyreactivity, including long third complementarity-determining regions of the heavy chain (HCDR3), are common among bNAbs but rare in naive, mature B cells (25, 26). This observation has led to speculation that bNAb polyreactivity may be a requisite trait for bNAbs (23).
Despite the potential importance of host mimicry by HIV-1 in mitigating protective immunity, there has been no systematic and quantitative assessment of poly- and autoreactivity among HIV bNAbs or, equally importantly, for the dominant nNAbs that commonly arise during HIV-1 infection. The few prior studies have mostly utilized clinical assays focused on diagnostic autoantigens and/or have not included significant numbers of nNAbs (17, 27, 28) as controls for general effects of infection, e.g., inflammation and chronic antigen (Ag) exposure.
In this study, we characterize the polyreactivity and autoreactivity of 22 HIV-1 bNAbs and 9 nNAbs in human protein microarrays that contain >9,400 full-length human proteins. Our characterizations reveal that poly- and autoreactivity are significantly more frequent in the bNAb group, both for individual Abs and Ab lineages. These observations suggest that these attributes are determined by the nature of specific neutralizing epitopes and are not generally attributable to HIV-1 infection. In addition, we identify novel host epitopes recognized by HIV-1 bNAbs and among these, human ubiquin-protein ligase 3A (UBE3A), was recognized by unrelated groups of CD4bs bNAbs. These data support the hypothesis that immunological tolerance hinders bNAb generation to the HIV-1 Env CD4bs as well as to the MPER (18, 19, 21, 29). We conclude that host mimicry constitutes a significant factor in limiting effective humoral immunity to HIV-1.
MATERIALS AND METHODS
HIV-1 Abs and Ags.
VRC01, VRC03, PG16, and 2G12 Abs used in this study were provided by the NIH AIDS reagent program. 2F5 and 4E10 were obtained from Polymun. NIH45-46 Ab was provided by Pamela Bjorkman, California Institute of Technology. VRC02 and VRC07 Abs were provided by John Mascola, NIH Vaccine Research Center. Additional bNAbs, namely, CH103, CH106, CH98, CH31, 10E8, PG9, CH01, CH03, PGT121, PGT125, PGT128, and PGT145 Abs, and nNAbs HG131, HG107, CH58, CH59, F39F, 19b, A32, 48d, and 17b were provided by the Duke University Human Vaccine Institute. All bNAbs originated from patients chronically infected with HIV-1 as were the F39F, 19b, A32, 48d, and 17b nNAbs. The HG131, HG107, CH58, and CH59 nNAbs were recovered from vaccinees in the RV144 vaccine trial (see Tables S1 and S2 in the supplemental material).
Recombinant HIV-1 gp140 (JR-FL [30]) and gp120 (CH505 T/F [7]) proteins were provided by the Duke Human Vaccine Institute for enzyme-linked immunosorbent assay (ELISA) to determine cross-inhibition by UBE3A.
Control Abs.
To control for Ab poly- and autoreactivity, we compared three isotype-matched human Abs, 151K (18), infliximab (Remicade; Centocor Ortho Biotech Inc.) (31), and palivizumab (Synagis; MedImmune) (32) to identify an optimal comparator. 151K is a human myeloma protein [IgG1(κ)] (catalog no. 0151K-01; SouthernBiotech), palivizumab [IgG1(κ)], and infliximab [IgG1(κ)] are therapeutic humanized Abs specific for the fusion protein of respiratory syncytial virus (RSV) and human tumor necrosis factor alpha (TNF-α), respectively (31, 32). 151K has been used previously as a protein microarray comparator for MPER bNAbs (18).
Protein array.
Abs were screened for binding on protein microarrays (ProtoArray) (catalog no. PAH0525101; Invitrogen) precoated with >9,400 human proteins in duplicate. The binding patterns of human bNAbs were compared to the human myeloma protein 151K in lot-matched arrays. Array-bound anti-human IgG served as the loading control for the detection Ab, and array-bound human IgG served as the loading control for the secondary reagent.
Abs were screened for reactive Ags on protein microarrays following the manufacturer's instructions and as described previously (18). Briefly, the ProtoArray microarray (Invitrogen) was blocked and incubated on ice with 2 μg/ml of HIV-1 Ab or isotype control 151K for 90 min. Ab binding to array protein was detected with 1 μg/ml of Alexa Fluor 647-labeled anti-human IgG (Invitrogen) secondary Ab. The ProtoArray microarrays were scanned using a GenePix 4000B scanner (Molecular Devices) at a wavelength of 635 nm, with 10-μm resolution, using 100% power and 650 gain. Fluorescence intensities were quantified with GenePix Pro 5.0 program (Molecular Devices) using lot-specific protein location information provided by the microarray manufacturer.
Protoarray data analysis.
The fluorescence intensity of HIV-1 Abs binding to each protein on the microarray was graphed against that of control Ab, 151K. The distance of each data point to the reference line, y = x (i.e., R = 1 [the diagonal line]), was determined using the distance formula: d = (x − y)/
To assess HIV-1 Ab polyreactivity, the distance of each data point to a reference line, y = x, was determined and graphed as a frequency histogram, with a mean fluorescence intensity (MFI) bin size of 0.02 (resolution threshold of GenePix 4000B scanner). The Gaussian mean of array protein distances from the y = x reference line is the polyreactivity index (PI), and determined by GraphPad software. When the PI of an HIV-1 Ab is greater than the PT (>0.21), the mean of the MFI for all array proteins was >2-fold over the control mean, defining the test Ab as polyreactive. Abs that were not polyreactive (PI ≤ 0.21) were defined as autoreactive if they recognized specific human proteins ≥500-fold more avidly than 151K.
Plasmids.
The ultimate open reading frame (ORF) clone in pENTR 221 vector containing the human UBE3A mRNA, transcript variant 2 (GenBank accession number NM_000462.3) (Invitrogen) was modified by the introduction of BamHI and NotI restriction sites. The BamHI/NotI UBE3A cleavage product was inserted into the pGEX-4T-1 vector (GE Healthcare). Truncation mutants of UBE3A were made by PCR, and the sequence was confirmed. Mutant UBE3AΔ700-875 was made with the following primers: 5′ GTTCAAGGACAGCAGTTGGCGGCCGCATCGTGACTG and 3′CAGTCACGATGCGGCCGCCAACTGCTGTCCTTGAAC. Mutant UBE3AΔ1-700 was made with the following primers: 5′ GATCTAAAGGAAAATGGTGCGGCCGCATCGTGACTG and 3′ CAGTCACGATGCGGCCGC ACCATTTTCCTTTAGATC. Mutant UBE3AΔ520-875 was made with the following primers: 5′ GTTCAAGGACAGCAGTTGGCGGCCGCATCGTGACTG and 3′ CAGTCACGATGCGGCCGCCAACTGCTGTCCTTGAAC. Mutant UBE3AΔ1-519 was made with the following primers: 5′ CTGGTTCCGCGTGGATCCAATCCATATTTGAGACTC and 3′GAGTCTCAAATATGGATTGGATCCACGCGGAACCAG.
Protein expression.
Glutathione S-transferase (GST)–UBE3A fusion protein and truncation mutants were expressed in Escherichia coli BL21 (New England BioLabs), and purified by glutathione-Sepharose affinity chromatography (GE Healthcare) following the manufacturer's instructions and as described previously (18). Briefly, E. coli BL21 (New England BioLabs) harboring pGEX4T1-UBE3A, pGEX4T1-UBE3AΔ520-875, pGEX4T1-UBE3AΔ1-519, or pGEX4T1 vector was grown at 37°C until it reached an optical density at 600 nm (OD600) of 0.4. Isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.4 mM (33). After 3-h induction at 37°C, cells were harvested by centrifugation at 6,000 × g for 15 min and resuspended in cold phosphate-buffered saline (PBS) with 1% protease inhibitor (Sigma-Aldrich). The cells were lysed with a probe sonicator and pelleted via centrifugation at 30,000 × g for 30 min. The GST-UBE3A fusion proteins were purified from lysate by glutathione-Sepharose affinity chromatography (GE Healthcare). Quantification of GST-UBE3A protein in lysate was done via SDS-PAGE and detection by Western blotting with anti-GST Ab (Sigma-Aldrich).
ELISA.
UBE3A binding by bNAbs was determined in a sandwich ELISA (18). Briefly, 96-well microplates (BD Biosciences) were coated with human bNAbs at 2 μg/ml overnight and blocked with permissive buffer (PBS, 0.5% bovine serum albumin [BSA], and 0.1% Tween 20) or stringent buffer (PBS, 4% whey protein, 15% normal goat serum, and 0.5% Tween 20). Recombinant UBE3A protein (Abcam) was added at 2 μg/ml for incubation in blocking buffer and 3-fold serially diluted. Mouse monoclonal anti-UBE3A Ab (2F6; Sigma-Aldrich) and polyclonal UBE3A-specific IgG (Thermo Fisher Scientific) were used to detect bound UBE3A. Secondary reagent goat anti-mouse IgG-HRP (affinity-purified and adsorbed goat serum IgG labeled with horseradish peroxidase [HRP]) (SouthernBiotech) was used to detect bound Ab. CD4 ELISA was performed similarly, with soluble CD4 (SinoBiological) coated at 2 μg/ml, and biotin anti-human CD4 (OKT4, BioLegend) used as positive control. For FAM84A (family with sequence similarity 84, member A) and PACRG (parkin coregulated gene protein homolog) ELISA, the plate was coated with 2 μg/ml proteins (Abnova), blocked with either permissive or stringent buffer, and detected by 10E8, 19b, and 151K serially diluted in blocking buffer.
Competitive inhibition ELISA with bNAbs was performed similarly with the following modifications. In human bNAb competition ELISA, VRC01 was coated onto ELISA plates at 2 μg/ml; after washing, HIV-1 JR-FL gp140 (2 μg/ml) and serial dilutions of competitor bNAbs or control Ab 151K (50 μM) were added and incubated for 1 h at ≈22°C. After incubation, the plates were washed, and the V4-SE Ab, a musinized 2F5 recombinant Ab (mouse Cγ2b and Cκ) specific for the HIV-1 gp41 MPER, was used to detect bound gp140; in turn, bound V4-SE was detected by the addition of anti-mouse IgG-HRP. When using UBE3A to inhibit bNAb binding to JR-FL gp140, JR-FL gp140 (1 μg/ml) was reacted with plate-bound VRC01 in the presence of increasing concentrations (0.5 nM to 1 μM) of recombinant UBE3A (heterologous inhibition) (Abcam) or HIV-1 gp120 (homologous inhibition). Bound gp140 was detected by the addition of V4-SE, followed by anti-mouse IgG-HRP.
Statistical analysis.
Significant differences in polyreactivity between bNAbs and nNAbs were tested using an exact Wilcoxon (Mann-Whitney) test, assuming two independent groups and without the assumption of normally distributed data. A supportive linear mixed-model regression, which places assumptions on the data such as normality, was performed to account for correlation among Abs from the same lineage. The results from the linear mixed model corroborated the results of the exact Wilcoxon test and have not been displayed. The exact binomial test was used to compare frequency of poly- and autoreactivity among bNAbs and nNAbs to the expected frequency (0.2 [24]). Statistical tests were performed using SAS 9.3.
RESULTS
Defining HIV-1 Ab binding patterns.
To assess polyreactivity and autoreactivity of HIV-1 bNAbs and nNAbs quantitatively, we compared the binding patterns of HIV-1 bNAbs and nNAbs to an isotype-matched (IgG1/κ) human myeloma protein (151K) in microarrays containing >9,400 full-length human proteins (18). The 151K Ab was chosen as an Ab binding control for consistency with previous work (18) and because it was not intentionally generated and has not been selected for the absence of poly- or autoreactivity. In contrast, therapeutic Abs cannot exhibit significant levels of off-target binding (31, 32). Nonetheless, compared to palivizumab, the vast majority of array proteins were bound equivalently by palivizumab and 151K (Fig. 1A), as evidenced by the tight distribution of data points along the diagonal binding axis. Thus, 151K and palivizumab, a therapeutic Ab, selected for little or no off-target binding, have comparably low levels of polyreactivity.

151K as a control for protein microarray. (A to C) Protein array binding by palivizumab (A), infliximab (B), and 151K (C). [151KMFI] is the MFI of 151K binding. (A and B) Axis values are MFI in palivizumab and infliximab array (x axis) or 151K array (y axis). (C) Pairwise comparison of duplicate proteins binding to 151K in the same array. Axis values are MFI in duplicate a (x axis) or duplicate b (y axis). Each dot represents one array protein replicate. A diagonal line indicates equal binding of two comparators. Internal controls for loading of Ab and secondary detection reagent were equally bound by Ab pairs (box). Dashed lines indicate the 500-fold signal/background ratio defined as the cutoff for significant autoreactivity. Circles identify protein ligands that bound ≥500-fold over the comparator.
As previously shown (18), 151K specifically bound one human protein, betaine-homocysteine methyltransferase 2 (BHMT2) >500-fold more avidly than it bound palivizumab (Fig. 1A). Palivizumab showed no evidence for autoreactivity (Fig. 1A). 151K is, therefore, an appropriate negative control for polyreactivity, and serves as a positive control to identify autoreactivity.
Analogous comparisons of 151K to infliximab also demonstrated that virtually all array proteins were bound equivalently by infliximab and 151K (Fig. 1B). As expected, infliximab specifically bound TNF-α >500-fold more avidly than it bound 151K (Fig. 1B); TNF-α was the only array protein bound by infliximab above the 500-fold threshold, suggesting that our criteria of >500-fold difference is stringent and conservative.
Internal controls for Ab loading, intracellular high-affinity IgG receptor TRIM21 (34), and anti-human IgG ensured that comparable amounts of Abs were present in all arrays; human IgG on the array served as a loading control for the secondary detection reagent (Fig. 1A and andB)B) (18).
To measure the experimental variability and dynamic range of 151K binding, we compared the MFI of 151K binding to duplicate proteins within the same microarray chip; these values were plotted against each other and consistently aligned along the diagonal axis with little variation, indicating high intraarray reproducibility (Fig. 1C). Ideally, differences in MFI intensity over all proteins in the array (as the mean displacement from the diagonal [151KaMFI = 151KbMFI]), i.e., no experimental or measurement variability, is 0; the observed intraarray value for five independent arrays was 0.001 (±0.004) (Fig. 1C).
Comparison of interarray MFI values was determined by comparing 151K binding to 151K binding in 5 independent arrays. In this comparison, the ideal value should be 0.001, the intraarray variability. The observed value, −0.008 (±0.086) (data not shown), documents the interarray variability of this assay.
We define HIV-1 Abs as polyreactive when the averaged MFI intensity over all array proteins are >2-fold above the matched 151K average. This >2-fold increase in binding is equivalent to a displacement of mean distance from the diagonal (151KMFI = HIV AbMFI) >0.21 (log10 2/
Polyreactive Abs were excluded from analyses for autoreactivity to minimize the roles of avid, but unspecific Ab-Ag interactions. Autoreactive Abs were defined as nonpolyreactive Abs that bound to array protein(s) with avidities ≥500-fold of the 151K control. This value was based on our earlier study of the 2F5 and 4E10 bNAbs; Ab-protein binding in microarrays with this signal-to-control ratio was always confirmed by stringent ELISA, whereas weaker avidities were not (18). A subset of polyreactive bNAbs bound selected array proteins ≥500-fold more avidly than the 151K control; nonetheless, in these cases, the bNAb subset was defined only as polyreactive.
Examples of these definitions are presented in Fig. 2 to to4.4. The CH98 bNAb was isolated from an HIV-1-infected individual with systemic lupus erythematosus (SLE), and is known to be polyreactive in AtheNA assay, HEp-2 immunofluorescence assay, and protein microarray (22). CH98 was therefore used as a positive control in our assay to validate the PI threshold of polyreactivity. In the protein microarray, CH98 reacted more avidly with ≈92.3% of the 9,400 microarray proteins than did the 151K control (Fig. 2A), exhibiting a PI value of 0.51 (Fig. 3A). This polyreactivity is readily apparent in the skewed distribution of 151KMFI/CH98MFI ratios, with most data points falling below the equivalence diagonal (Fig. 2A). A histogram of this displacement illustrates a significant shift (>0.21) from the equivalence diagonal (Fig. 3A). In contrast to the arrayed proteins, internal loading controls (Fig. 2A) were bound equally by CH98 and 151K, eliminating the possibility that the skewed Ag binding pattern by CH98 was an artifact of unequal Ab loading.
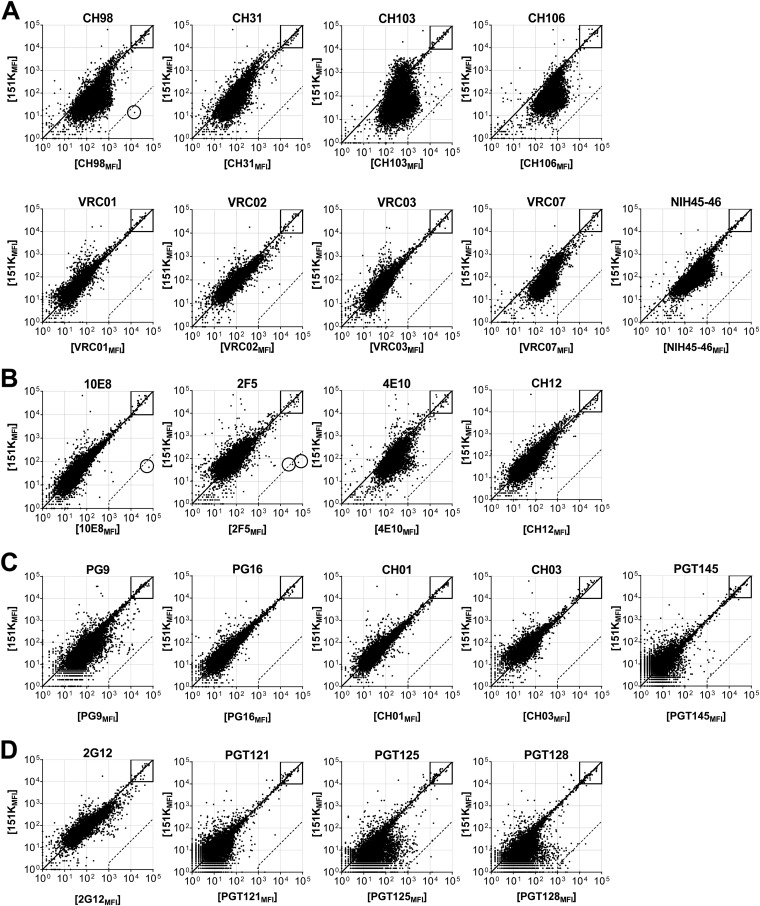
Protein microarray binding by bNAbs. (A to D) Representative ProtoArray summary for protein arrays blotted with CD4bs bNAbs (A), MPER bNAbs (B), V1/V2 bNAbs (C), and V3-glycan bNAb (D), or 151K control. Axis values are relative fluorescence signal intensity in the 151K array (y axis) or test Ab array (x axis). Each dot represents the average of duplicate array proteins. A diagonal line indicates equal binding by test Ab and 151K. Internal controls for loading of Ab and secondary detection reagent were equally bound by Ab pairs (boxes). Dashed lines indicate the 500-fold signal/background ratio defined as the cutoff for autoreactivity. Circles identify protein autoligands recognized by autoreactive bNAbs.
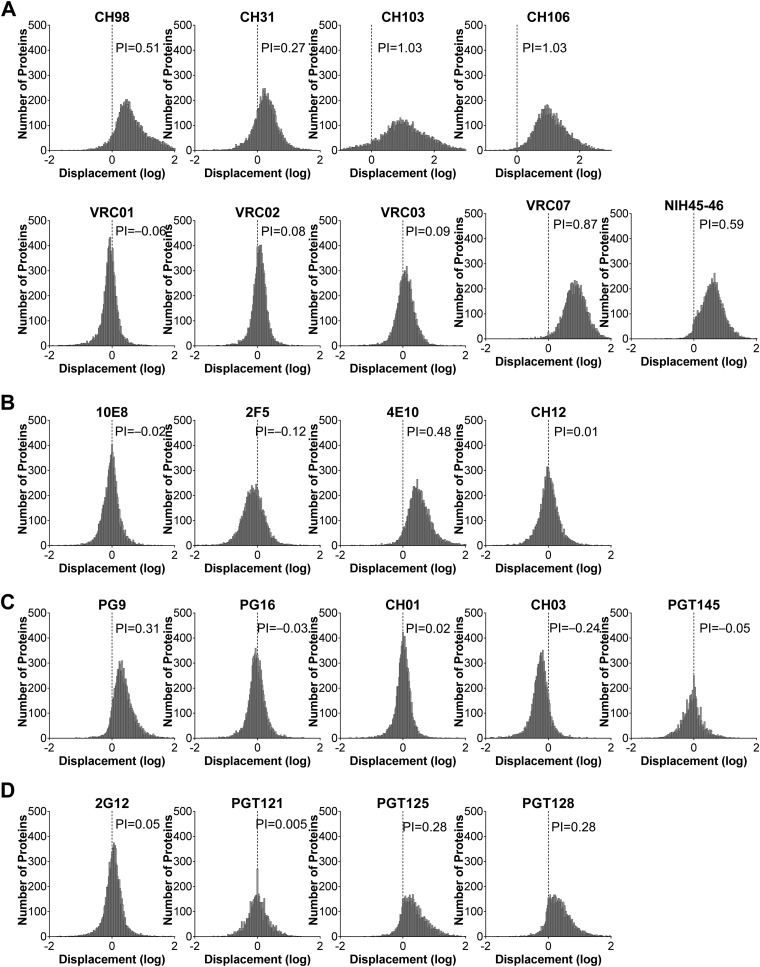
Polyreactivity indices of bNAbs. (A to D) Frequency histogram of protein displacement (log) from the diagonal line by bNAbs specific for CD4bs (A), MPER (B), V1/V2 (C), and V3-glycans (D), compared to isotype control 151K in Fig. 2. Negative displacements indicate stronger binding by control 151K. The Gaussian mean of all array protein displacements is termed the polyreactivity index (PI). A PI value of 0.21 suggests 2-fold-stronger overall binding by test Ab than control 151K, and it was defined as the threshold of polyreactivity.
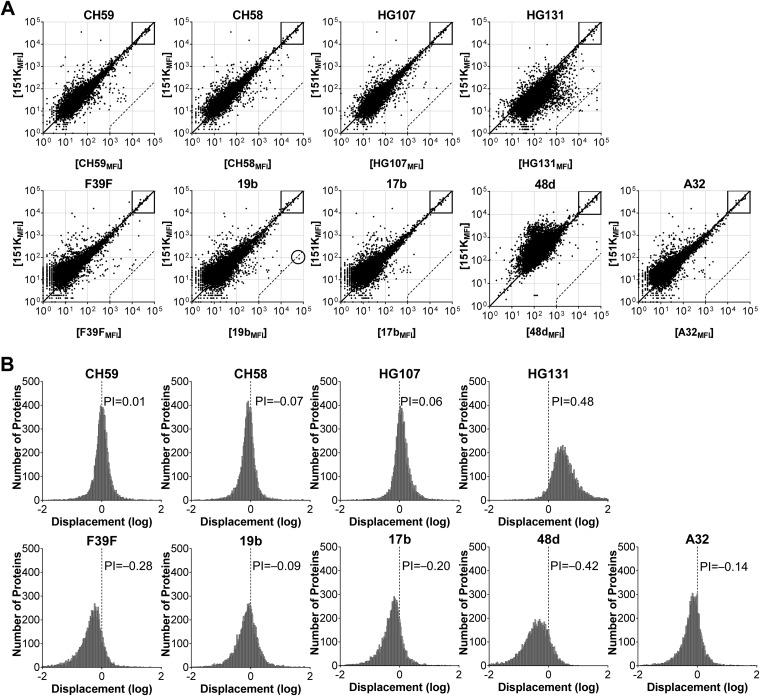
Protein microarray binding and polyreactivity indices of nNAbs. (A and B) Representative ProtoArray binding fluorescence intensity (A) and frequency histogram (B) of displacement (log) from the diagonal line in panel A by HIV-1 nNAbs compared to the 151K control. (A) Each dot represents an individual protein, and axis values are the means of duplicate fluorescence intensities in the control array (y axis) and nNAb array (x axis). Boxes denote internal controls for loading of Ab and secondary detection agent. Dashed lines are the cutoff autoreactivity, as defined by the 500-fold nNAbMFI/151KMFI ratio. Circles highlight autoantigens by nonpolyreactive nNAbs. (B) Displacement of each protein from the diagonal in panel A was determined and graphed in a frequency graph with a bin size of 0.02. Positive displacement indicates stronger binding by nNAb than 151K and vice versa. PI is the Gaussian mean of all displacement values. A PI value of 0.21 suggests 2-fold-stronger overall binding by test nNAb than control 151K, and it was defined as the threshold of polyreactivity.
CH98 also bound to a single human protein, STIP1 homology and U-box containing protein 1 (STUB1) (Fig. 2A) >500-fold more avidly than it bound to 151K (Fig. 2A), meeting the definition for significant autoreactivity. However, because bNAb CH98 is polyreactive, it is not identified as autoreactive.
The CH59 nNAb (35) bound the substantial majority of arrayed proteins with avidity similar to that of the 151K control (Fig. 4A). Indeed, the displacement histogram exhibited a normal distribution with a mean value of 0.01 (Fig. 4B), indicating that the CH59 nNAb was neither more nor less polyreactive than 151K Ab. CH59 did not bind to any array ligand with an avidity ≥500-fold greater than that of the 151K Ab (Fig. 4A). In consequence, we define CH59 to be neither poly- nor autoreactive; CH59 is highly specific for the V2 loop region of HIV-1 Env (35).
Finally, the MPER bNAb 10E8 (10) was not polyreactive compared to the 151K control (Fig. 2B), with a mean histogram displacement of −0.02 (Fig. 3B). Despite the similar overall binding pattern to 151K, 10E8 interacted with a single human protein, FAM84A (family with sequence similarity 84, member A) (Fig. 2B), with avidity ~1,150-fold greater than 151K (Fig. 2B). The 10E8 bNAb is defined as autoreactive.
Polyreactivity in HIV-1 bNAbs and nNAbs.
The cohort of 22 bNAbs tested are thought to represent 14 clonal lineages and cover the four major neutralization epitopes (25). The VRC01 family (VRC01, VRC02, VRC03, VRC07, and NIH45-46), CH98, CH30-34 (CH31), and CH103-106 (CH103 and CH106) lineages recognize CD4bs. 2F5, 4E10, 10E8, and CH12 lineages bind MPER. PG9/PG16 (PG9, PG16), CH01-04 (CH01 and CH03) and PGT141-145 (PGT145) lineages map to the V1V2-glycan epitopes. 2G12, PGT121-123 (PGT121), and PGT125-131 (PGT125, PGT128) lineages recognize V3-glycans (Fig. 2 and and3;3; see Table S1 in the supplemental material). The nine nNAbs tested represent distinct lineages and are specific for HIV-1 Env V2 and V3 loop epitopes (CH58, CH59, HG107, HG131, F39F, and 19b) and CD4-induced (CD4i) gp120 epitopes (A32, 17b, and 48d) (Fig. 4; see Table S2 in the supplemental material). These nNAbs include both strain-specific (autologous) neutralizing Abs and nonneutralizing Abs; all are categorized as nNAbs.
In this way, 45% (10/22) bNAbs (VRC07, NIH45-46, CH103, CH106, CH98, CH31, 4E10, PG9, PGT125, and PGT128) were found to be polyreactive (Fig. 5A), with significant shifts in the mean of displacement in histogram plots of relative MFI (PI > 0.21) (Fig. 2 and and3).3). The remaining 12 bNAbs (VRC01, VRC02, VRC03, 2F5, 10E8, CH12, PG16, CH01, CH03, 2G12, PGT121, and PGT145) bound most array proteins with avidities similar to or less than that of the 151K control (PI ≤ 0.21) and were defined as nonpolyreactive (Fig. 2 and and33).

bNAbs are more polyreactive than nNAbs. (A) Summary of polyreactivity profile of 22 bNAbs and nine nNAbs as assessed in Fig. 3 and and4.4. Polyreactive indicates that the PI was >0.21. (B) Histogram of PI value distribution among bNAbs or nNAbs, with a bin size of 0.1. Gaussian best-fit regressions were imposed over the histogram.
In contrast, only a single (1/9 [11%]) nNAb (HG131) was found to be polyreactive. HG131 was derived from a vaccinee (RV144 ALVAC prime, AIDSVAX B/E protein boost) and binds to the gp120 V2 (M. Bonsignori, unpublished data). The remaining eight nNAbs (CH58, CH59, HG107, F39F, 19b, A32, 17b, and 48d) exhibited comparable or lower polyreactivity than that of the 151K Ab (Fig. 4).
As a group, the PI values for bNAbs (n = 22) were significantly higher than for nNAbs (n = 9) (P = 0.009 by exact Wilcoxon test) (Fig. 5B), indicating that bNAbs are generally more frequently polyreactive or more avidly polyreactive than are nNAbs. Indeed, nNAbs exhibited an expected Gaussian distribution of PI values, with a mean approximating 0 (i.e., equivalent overall binding to the 151K control). In contrast, the PI values of bNAbs exhibited a mixed distribution pattern and were enriched for high PI values (Fig. 5B).
As a group, bNAbs are more mutated than nNAbs (see Tables S1 and S2 in the supplemental material); these 22 bNAbs exhibit an average VH mutation frequency of 20.5%, whereas VH mutation frequencies in the nine nNAbs average 10%. However, the increased polyreactivity among bNAbs is not a direct result of higher mutation frequencies, as no correlation could be drawn between the PI value and VH mutation frequency among bNAbs and nNAbs (both separately and combined [unpublished data]). In addition, even among bNAbs and nNAbs with high mutation frequencies (>10%), PI values are not correlated to mutation frequency.
Polyreactivity of bNAb and nNAb lineages.
We next analyzed bNAbs grouped by clonal lineages, as Abs within lineages are clonally related and do not represent independent samples. As expected, clonally related bNAbs often exhibited similar patterns of polyreactivity, e.g., CH103 and CH106, PGT125 and PGT128, and CH01 and CH03 (Fig. 2 and and3).3). Two bNAb lineages, however, comprised significantly different binding patterns and PI values. First, the V1/V2 bNAb PG9 is polyreactive (PI = 0.31), whereas its sister clone, PG16, was not (PI = −0.03) (Fig. 2 and and3).3). Second, the VRC01 family bNAbs cluster into two binding groups; VRC01, VRC02, and VRC03 are not polyreactive (PI = −0.06 to 0.09), whereas VRC07 and NIH45-46 are (PI = 0.87 and 0.59, respectively) (Fig. 2 and and3).3). Both VRC07 and NIH45-46 share a 12-nucleotide HCDR3 insertion (36, 37) that appears to confer increases in both polyreactivity and neutralization potency (Fig. 2 and and3)3) (36, 37).
To determine whether the bNAb lineages are more polyreactive than nNAb, the mean PI value of bNAbs within the same clonal lineage was compared to that of the nNAbs. The lineage means of the bNAbs (n = 14) remain significantly higher than those of the nNAbs (n = 9) (P = 0.026 by exact Wilcoxon test). bNAb lineages are significantly more polyreactive (or enriched for polyreactivity) than nNAbs.
Whereas all bNAbs originated from patients chronically infected with HIV-1, nNAbs arose from both infected donors (F39F, 19b, A32, 48d, and 17b) and vaccinees (HG131, HG107, CH58, and CH59). The most rigorous comparison of polyreactivity is between bNAbs and nNAbs recovered from chronically infected patients. Restricting our analysis to nNAbs derived from infected donors (n = 5) still showed significantly less polyreactivity than that of bNAbs. This was true both for individual Abs (n = 22) (P = 0.0003 by exact Wilcoxon test) and clonal lineages (n = 14) (P = 0.0007 by exact Wilcoxon test).
Autoreactivity in HIV-1 bNAbs and nNAbs.
Of the 12 nonpolyreactive bNAbs, three (VRC01, 10E8, and 2F5) were found to react strongly (≥500-fold over 151K) with specific human proteins (3/12 [25%]) (Fig. 6A) (18), UBE3A isoforms 2 and 3 (VRC01), FAM84A (10E8), and KYNU and CKLF-like MARVEL transmembrane domain containing 3 (CMTM3) (2F5) (18). In contrast, only a single nonpolyreactive nNAb (1/8 [13%]), 19b, reacted significantly with an array protein. The 19b nNAb that is specific for the gp120 V3 loop (38) also bound to parkin coregulated gene protein homolog (PACRG) (Fig. 6A).
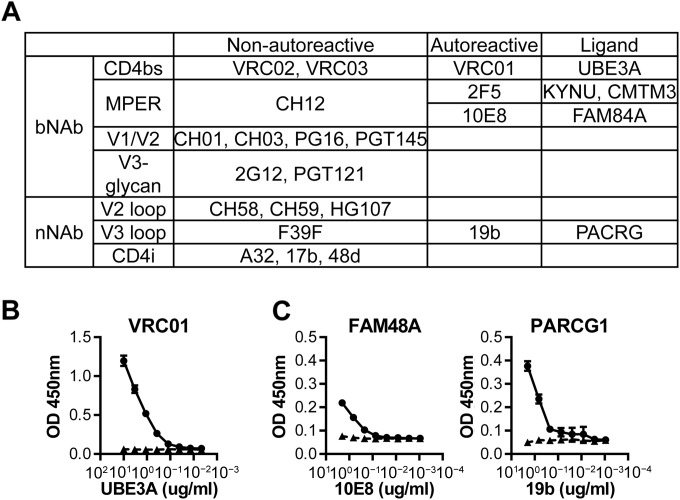
bNAbs are more autoreactive than nNAbs. Autoreactive human protein ligands were identified for nonpolyreactive bNAbs and nNAbs and confirmed for binding in ELISA under stringent conditions. (A) Summary of autoreactivity among nonpolyreactive bNAbs and nNAbs. Autoreactivity was defined as binding ≥500-fold stronger than 151K in protein array (Fig. 2 and and4,4, dashed lines). (B) VRC01 (circles) and 151K (triangles) were immobilized on ELISA plates, blocked under stringent conditions, and added with serially diluted UBE3A. Monoclonal Ab against UBE3A detects specific binding. BSA was used as a negative control. (C) FAM48A and PARCG1 were immobilized, blocked under stringent conditions, and detected with serially diluted 10E8, 19b (circles), and 151K (triangles). (B and C) Binding by all HIV-1 Abs were significant and specific under permissive ELISA conditions (unpublished data). Axis values are optical densities (OD) (y axis) or protein concentrations (μg/ml) (x axis). All experiments were repeated in two or more independent experiments. Error bars indicate standard deviations (SDs).
To confirm bNAb and nNAb reactivity with these array proteins, we obtained full-length recombinant human UBE3A, FAM84A, and PACRG proteins and tested the binding of VRC01, 10E8, and 19b in stringent ELISA conditions (18) (Fig. 6B and andC).C). Two types of ELISA were used, Ag capture (VRC01 [Fig. 6B]) and direct binding (10E8 and 19b [Fig. 6C]). Ag capture is considerably more sensitive than direct-binding ELISA. Binding to array proteins by VRC01, 10E8, and 19b were all preserved in stringent ELISA conditions (Fig. 6B and andCC).
UBE3A is bound by unrelated CD4bs bNAbs.
Four CD4bs bNAbs from two distinct clonal lineages, the VRC01 and VRC02 bNAbs and CH106 and CH103 bNAbs, bound human UBE3A more avidly than the 151K control Ab did (Fig. 7A). Only VRC01 bound UBE3A ≥500-fold above the 151K control (Fig. 7A) with the related VRC01 and VRC02 bNAbs exhibiting signal-to-control ratios of ≈800- and 400-fold, respectively. CH106 and CH103 bound UBE3A only ≈100-fold more avidly than 151K did, indicating substantially weaker interactions with UBE3A. Nonetheless, the binding of all four bNAbs to recombinant UBE3A protein was confirmed in a stringent ELISA (Fig. 7B) (18). In the ELISA, VRC01 and VRC02 exhibited much stronger binding to UBE3A than did CH106 and CH103 (VRC01 > VRC02 > CH106 > CH103) with the related CH103 and CH106 bNAbs also exhibiting higher background binding, likely due to their polyreactivity (Fig. 7A). Surface plasmon resonance estimates of UBE3A-VRC01 binding strength suggested a Kd (dissociation constant) of ≈30 nM (unpublished data). Such avidity is well within the range of autoreactive human Abs derived from autoimmune patients (9.1 nM to 58 μM [39]), suggesting that in vivo, VRC01 could be subject to tolerance control.
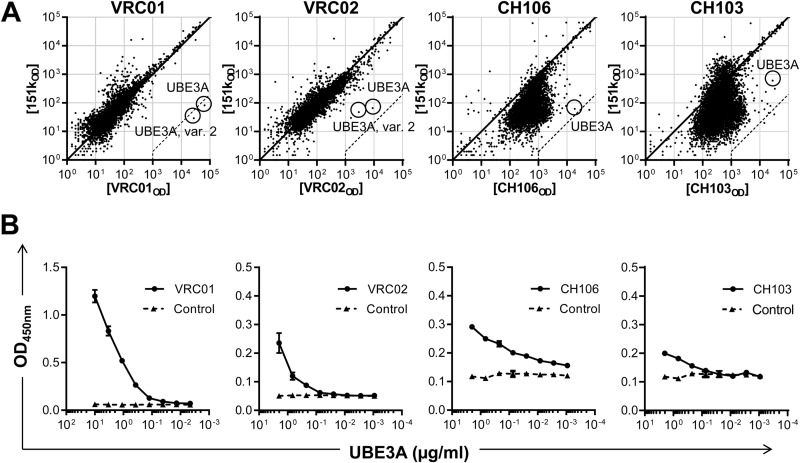
UBE3A is a shared autoantigen by four CD4bs bNAbs. UBE3A was recognized by VRC01, VRC02, CH106, and CH103 with different strengths in the protein array and ELISA. (A) Representative protein array summary for protein arrays blotted with CD4bs bNAbs VRC01, VRC02, CH106, CH103, or 151K control. Axis values are fluorescence intensity in the 151K array (y axis) or bNAb array (x axis). Each dot represents the average of duplicate proteins. A diagonal line indicates equal binding by test Ab and 151K. The dashed line indicates 500-fold MFI difference and serves as the cutoff for high-affinity autoreactivity. UBE3A proteins were marked with circles. (B) CD4bs bNAbs (circles) and 151K (triangles) were tested for binding to UBE3A in sandwich ELISA under stringent conditions as described in Materials and Methods. All experiments were independently repeated at least twice. Error bars reflect standard deviations (SDs).
To determine whether VRC01, VRC02, CH106, and CH103 recognize a common UBE3A epitope, we measured the inhibition of VRC01-UBE3A binding in ELISA by VRC01 (homologous inhibition) or by VRC02, CH106, or CH103 (heterologous inhibition) (Fig. 8A). Addition of VRC02 inhibited VRC01-UBE3A binding as well as VRC01 itself; these clonally related bNAbs bind to the same UBE3A determinant. CH106 and CH103, on the other hand, did not inhibit VRC01-UBE3A binding (Fig. 8A). Reciprocally, CH103, and less potently VRC01, inhibited the CH106-UBE3A binding, although not as well as homologous inhibition by CH106 (Fig. 8B). The incomplete but dose-dependent inhibition of CH106-UBE3A binding by CH103 may be the result of its lower affinity for UBE3A (Fig. 7B). Partial inhibition of CH106-UBE3A binding by VRC01 implies that the VRC02/VRC02 and CH103/CH106 epitopes on UBE3A are distinct but overlapping. This analysis is consistent with maps of the VRC01/VRC02 and CH106/CH103 epitopes on HIV-1 gp120 that posit distinct but adjacent determinants on Env (see Fig. S1 in the supplemental material) (7).
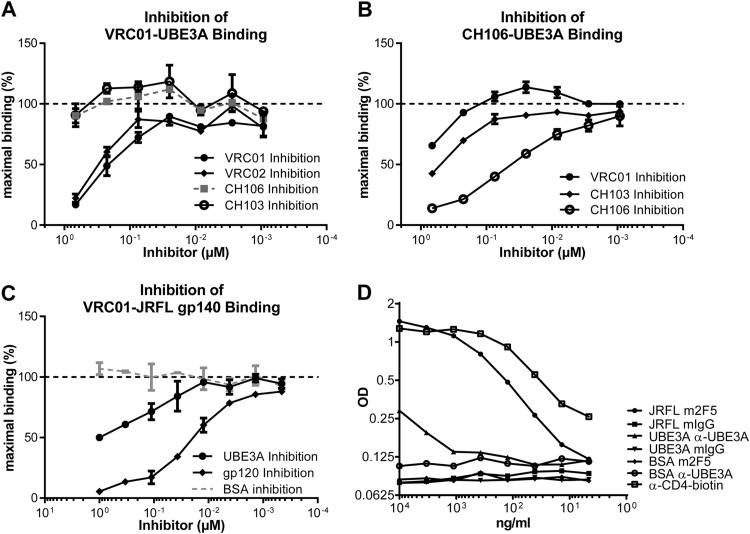
VRC01 binds UBE3A and CD4bs of gp120 in similar manners. (A to C) Inhibition of VRC01 binding to UBE3A (A), or JR-FL gp140 (C), or CH106 binding to UBE3A (B) assessed in ELISA as described in Materials and Methods. The y axis indicates OD percentage of maximal binding, which is determined as the mean reading without inhibitors (dashed line). The x axis depicts the molar concentration of inhibitor bNAb. Error bars indicate standard deviations. (D) Serially diluted JR-FL gp140, UBE3A, or BSA was added to an ELISA plate immobilized with human CD4. Then mouse 2F5, anti-UBE3A (α-UBE3A), or isotype control was added to detect specific binding. An HRP-conjugated Ab specific for mouse IgG was used to detect bound interactions. A biotinylated anti-CD4 and streptavidin-HRP served as positive control (empty squares). CD4 binding to gp140 is detected by mouse 2F5 and serves as positive control (filled circles).
To determine whether gp120 and UBE3A bind to VRC01 at a common site, we inhibited VRC01-gp140 binding by the addition of gp120 (homologous) or UBE3A (heterologous) inhibitor. VRC01 binding to the JR-FL gp140 was strongly inhibited by the introduction of CH505 transmitted/founder (T/F) gp120 (7) but also by the addition of UBE3A (Fig. 8C). Whereas homologous inhibition was virtually complete at 1 μM gp120, inhibition by UBE3A at the same concentration was only 50%. This inhibition was not due to increased protein concentrations, as the addition of BSA had no effect on VRC01-gp140 binding (Fig. 8C). The ability of UBE3A to inhibit VRC01-gp140 binding specifically and in a dose-dependent fashion indicates that VRC01 interacts with both ligands at a common paratopic site.
If gp120 and UBE3A share a epitope that interacts with the VRC01 paratope, it follows that UBE3A, like gp120, may be capable of binding CD4. Consequently, we tested UBE3A for binding to human CD4 coated on ELISA plates (Fig. 8D). Bound CD4 was recognized by OKT4, a CD-4-specific monoclonal antibody (MAb) and JR-FL gp140. Remarkably, UBE3A exhibited weak but specific binding to plate-bound CD4 (Fig. 8D). This result offers direct evidence that the epitope shared by UBE3A and HIV-1 gp120 overlaps with the structural motif recognized by both VRC01 and human CD4.
VRC01 binds UBE3A via a conformational epitope.
UBE3A, an E3 ubiquitin-protein ligase, is an ancient protein that is present in all bilaterian species (40). The ligase consists of two domains, an N-terminal substrate-binding domain and a C-terminal catalytic region common to all HECT E3 ubiquitin-protein ligases (40). UBE3A sequence is highly conserved among mammalian species with primates, rodents, and marsupials sharing >98% sequence identity (NCBI HomoloGene, unpublished data).
No significant amino acid sequence similarities were identified between UBE3A and gp120 (UniProt sequence alignments; unpublished data) indicating that the shared epitope is discontinuous; this finding is unsurprising, as HIV-1 gp120 interacts with VRC01 via a discontinuous, conformational epitope (41). Structural modeling of VRC01-UBE3A binding is hindered by the absence of a high-resolution structure for the complete UBE3A molecule. Only the catalytic, C-terminal HECT domain of human UBE3A has been resolved (42). That the HECT domain is conserved among 20 human E3 ubiquitin ligases (40) suggests that the VRC01 epitope is not located in this C-terminal domain, as other HECT domain-containing E3 ubiquitin ligases (UBE3C, HERC3, WWP1, WWP2, and HUWE1 [40]) were not recognized by VRC01 in the protein microarray (Fig. 7A). In contrast, the N termini of E3 ubiquitin protein ligases that determine substrate specificity vary substantially from one another (42), and we hypothesize that this N-terminal domain contains some or all of the UBE3A epitope bound by the VRC01 bNAb.
In an attempt to locate the VRC01 binding site on UBE3A, we constructed seven truncation mutants (T1 to T7) of human UBE3A that were expressed in bacteria (Fig. 9). These mutants represented serial truncations of the UBE3A N-terminal domains (n = 5) or C-terminal domains (n = 2) and an amino-terminal GST tag. All complete and truncated proteins were purified over glutathione columns and analyzed for reactivity against UBE3A-specific rabbit IgG, the UBE3A-specific 2F6 MAb, and the VRC01 bNAb. All three Abs bound the full-length GST-UBE3A protein, and all the truncation mutants were recognized by the polyclonal UBE3A Ab (Fig. 9). The 2F6 MAb is known to react with an epitope in the N-terminal, substrate-binding domain of UBE3A; 2F6 MAb reacted with all UBE3A truncation mutants except T2 (deletion of the N-terminal domain) and T6 (deletion of first 400 residues of the N-terminal domain), locating the 2F6 epitope between residues 300 and 400 in the substrate-binding domain. The binding of polyclonal IgG and the 2F6 MAb to the mutant UBE3A proteins rules out a general loss of native structure. None of the truncation mutants, however, retained VRC01 binding. We conclude that an easily disturbed structural motif defines the VRC01 epitope on UBE3A.
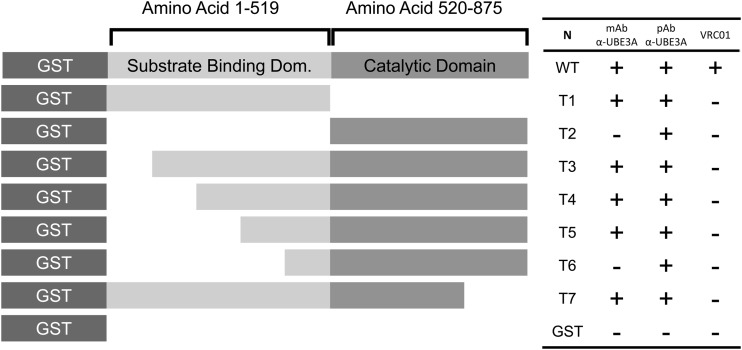
VRC01 recognizes UBE3A via a conformational epitope. Truncated (T1 to T7) and full-length (WT) UBE3A proteins were synthesized with an N-terminal GST tag or with a GST control and tested for binding in an ELISA to VRC01, polyclonal, or monoclonal anti-UBE3A (α-UBE3A) as detailed in Materials and Methods. Symbols: +, binding; −; no binding. N, name; WT, wild type.
DISCUSSION
It is generally acknowledged that HIV-1 bNAbs capable of neutralizing a broad range of viral isolates are often autoreactive or polyreactive (1, 25). HIV-1 bNAbs possess one or more unusual structural characteristics, including very long HCDR3s and/or extraordinary frequencies of V(D)J mutations (1, 25), that are correlated with Ab polyreactivity, and it is possible that these structural properties, and poly- or autoreactivity, are necessary for bNAb activity. This heightened self-reactivity could be, however, an epiphenomenon of B-cell selection during chronic HIV-1 infection (43), as persistent inflammation promotes the accumulation of poly- and autoreactive Abs (22, 44). Consequently, our study characterizes both bNAbs and nNAbs so as to determine whether polyreactivity/autoreactivity (poly/autoreactivity) are the products of the infection milieu (e.g., chronic inflammation, prolonged Ag exposure, CD4 T-cell depletion, etc.) or is linked specifically to bNAb activity.
Poly- and autoreactivity of bNAbs have been more commonly assessed by ELISA (cardiolipin, double-stranded DNA [dsDNA], and single-stranded DNA [ssDNA]), cell immunofluorescence assay (HEp-2), and AtheNA assay (cell nucleus antigens) (1). Such assays are useful but potentially limited either by the number of autoantigens tested (ELISA and AtheNA assay) or by preparation artifacts, interference, variable levels of antigen expression, and nonquantitative assessments of reactivity. In contrast, we determined the specificities of HIV-1 Abs in standardized protein microarrays containing >9,400 purified human recombinant proteins. Binding by bNAbs and nNAbs were compared to a single isotype-matched control myeloma Ab, 151K, that arose spontaneously and has not been selected with regard to paratopic specificity (18). Comparisons of 151K to two therapeutic Abs, palivizumab and infliximab, demonstrate that 151K has little or no polyreactivity but does bind avidly to BHMT2 (Fig. 1A and andB),B), a human methyltransferase that is abundant in kidney and liver (BioGPS, probe set 219902). These dual properties, little polyreactivity and an avid and specific autoreactivity, make 151K an ideal control for these studies. In addition, 151K was used previously in a study of two MPER bNAbs, and its use here ensures that both studies are directly comparable. By our definitions (Materials and Methods), 10 of the 22 (45%) HIV-1 bNAbs tested were polyreactive, in contrast to only one (1/9 [11%]) HIV-1 nNAb (Fig. 5A). This difference in polyreactivity between the bNAb and nNAb groups is statistically significant (P = 0.009 by exact Wilcoxon test). Although small sample sizes precluded statistical comparisons among different bNAb types, at least one polyreactive bNAb was present in each of the four major bNAb classes: CD4bs, MPER, V1/V2, and V3-glycan (Fig. 5A). bNAb polyreactivity is not restricted to the MPER bNAb class.
We defined autoreactivity only in Abs that were not polyreactive because of the difficulty in assessing specific binding in the presence of strong nonspecific interactions. The threshold for autoreactivity, an MFI ≥500-fold over the MFI of the 151K control, was chosen empirically to be consistent with previous work (18) and, more significantly, the results of V(D)J knock-in animal studies (19,–21). We have now identified three nonpolyreactive bNAbs, VRC01, 10E8, and 2F5, as significantly autoreactive. These bNAbs avidly bind UBE3A, FAM84A, and KYNU/CMTM3, respectively (Fig. 6). In addition, the nNAb 19b was shown to recognize PACRG (Fig. 6). Taken together, 13/22 of bNAbs were either poly- or autoreactive (59%), whereas only 2/9 of nNAb exhibited poly- or autoreactivity (22%). Given that approximately 20% of mature naive B cells in healthy humans are poly- or autoreactive (24), the frequency of poly/autoreactivity among nNAbs is unexceptional (exact binomial test H0: probability of autoreactivity/polyreactivity [prob(auto/poly)] = 0.2, P = 0.88]. In contrast, the bNAb cohort studied was significantly enriched for poly/autoreactivity [exact binomial test H0: prob(auto/poly) = 0.2, P < 0.0001].
Importantly, the nNAbs derived from infected patients (n = 5) (F39F, 19b, A32, 17b, and 48d) are significantly less polyreactive than nNAbs derived from vaccinated patients (n = 4) (CH58, CH59, HG107, and HG131) (P = 0.016 by exact Wilcoxon test). Thus, bNAbs are significantly more polyreactive than nNAbs that originated from only infected donors (n = 5) (individual bNAbs [n = 22, P = 0.0003]; bNAb lineages [n = 14; P = 0.0007]). This result suggests that despite their similar origins in infected patients, poly- and autoreactivity appear to be associated only with HIV-1 bNAbs (17, 23, 28, 45).
Poly- and autoreactivity in bNAbs is not the inevitable result of high V(D)J mutation frequencies and/or long HCDR3 sequences. For example, even though a third of V(D)J nucleotides encoding the variable regions of the VRC03 bNAb are somatic mutants (5) and whereas the HCDR3 of bNAb PG16 comprises a remarkable 84 nucleotides (4), neither VRC03 nor PG16 is auto- or polyreactive (Fig. 2 and and3).3). Indeed, we found no significant correlation between the PI values and mutation frequencies among bNAbs and nNAbs (both separately and combined [unpublished data]).
Our definitions of poly- and autoreactivity were empirically determined and chosen to be conservative. We note that B-cell development is strongly blocked in knock-in mice expressing the 2F5 VDJ heavy-chain (H-chain) rearrangement in association with the complete repertoire of endogenous light chains (L-chains) even though the affinity for the canonical 2F5 ligand is substantially reduced (19). Consistently, 2F5 binds KYNU and CMTM3 in the array >500-fold more avidly than it binds 151K (Fig. 2B). In addition, 151K, but not palivizumab, reacts with BHMT2 >500-fold more avidly than its comparator, an avidity comparable to that of infliximab for TNF-α (Fig. 1A and andB).B). The 500-fold threshold for specific autoreactivity is, therefore, consistent with high-avidity, functional binding. We caution that this threshold may be too strict. Indeed, binding of VRC02 to UBE3A under stringent ELISA conditions was strong, even though the reaction in microarrays did not meet the 500-fold cutoff (Fig. 7).
Similarly, the overall twofold-stronger binding (PI > 0.21) definition for polyreactivity is chosen to be conservative and in concert with our characterization of the 4E10 bNAb (18). Knock-in mice expressing the 4E10 V(D)J rearrangements exhibit impaired B-cell development (21, 46). However, even if a higher PI threshold were chosen (e.g., 0.27, or 2.5-fold-stronger overall binding), the frequency of polyreactive bNAbs and nNAbs would not have changed (Fig. 3 and and4).4). The final measure of significant poly- and autoreactivity is the generation of V(D)J knock-in mice (19,–21, 46,–48), and additional knock-in strains will be necessary to determine the significance of off-target bNAb reactivities. However, if the 2F5 and 4E10 mice are reasonable guides, we expect that most, if not all, of the poly- and autoreactive bNAbs studied here will be subject to some degree of immunological control. Regardless, the significantly higher PI values of bNAbs than nNAbs demonstrate that as a group, bNAbs exhibit polyreactivity as a fundamental characteristic.
The binding of UBE3A by unrelated CD4bs bNAb groups is intriguing. UBE3A, an E3 ubiquitin-protein ligase, is abundant in many human tissues (49), and it expresses an HIV-1 gp120 epitope that overlaps the CD4bs determinants recognized by VRC01, VRC02, CH106, and CH103 (Fig. 7 and and8).8). Interestingly, bNAb avidity for UBE3A correlates well with neutralization breadth, in the order of VRC01 > VRC02 > CH106 > CH103 (see Table S1 in the supplemental material). The four bNAbs reactive with UBE3A are clonally related pairs: VRC01 and VRC02 were derived from the same patient, and CH103 and CH106 were derived from a different donor (5, 7). Although both CH106/CH103 and VRC01/VRC02 bind the CD4bs of HIV-1 gp120, the bNAb pairs approach the CD4bs from different angles and interact distinctly with epitopic residues (7, 41). We demonstrate here that VRC01 and CH106 recognize UBE3A via overlapping epitopes (Fig. 8A and andB).B). Further, VRC01 binds UBE3A and CD4bs of gp120 at a common paratopic site (Fig. 8C). Remarkably, UBE3A binds specifically, albeit weakly, to CD4 (Fig. 8D); we conclude that UBE3A contains a structural motif shared with HIV-1 gp120 that mimics the VRC01/CH106 epitopes and CD4bs. That the UBE3A epitope mimics the CD4bs of HIV-1 implies tolerizing pressure for B cells expressing B-cell receptors (BCR) similar to those that define the VRC01/02 and CH106/103 lineages.
UBE3A is highly conserved in mammalian species; indeed, the amino acid sequences of human, chimpanzee, macaque, and baboon UBE3A are 100% identical (NCBI HomoloGene), suggesting that generation of CD4bs bNAbs in all primates may be restricted by the shared UBE3A-CD4bs epitope. Interestingly, both KYNU and UBE3A, conserved mammalian proteins mimicked by HIV-1 neutralizing epitopes, are crucial components of tryptophan metabolism. KYNU, the enzyme that is mimicked by the 2F5 epitope of HIV-1 MPER (18), mediates cleavage of l-kynurenine into anthranilic acid (50), and UBE3A is involved in converting 5-hydroxyindoleacetate to 5-hydroxyindoleacetylglycine (51). That two major HIV-1 bNAb epitopes mimic human proteins of the same pathway is intriguing, as tryptophan metabolites have been implicated in regulating both innate and adaptive immune activation (52), as well as neurological functions (53). Indeed, hypomorphic mutations in the UBE3A gene cause Angelman syndrome (54), a neurologic disorder; quinolinate, a kynurenine pathway product, has been implicated in AIDS-related dementia (55).
The generation of the MPER bNAbs 2F5 and 4E10 is controlled by immunological tolerance (18,–21, 46) and our identification of UBE3A as another self-antigen recognized by HIV-1 bNAbs provides a new example of host mimicry by HIV-1. Such mimicry conceals vulnerable neutralization sites by hiding in plain sight: immunological tolerance purges the primary immune response of those lymphocytes most suited for protective immunity. This method of immune evasion can be highly effective (18, 56). We hypothesize that host mimicry by pathogens is evolutionarily advantageous and is more widespread than generally appreciated (57).
The high frequencies of poly- and autoreactivity (Fig. 5 and and6)6) among HIV-1 bNAbs are consistent with our hypothesis that HIV-1 evades protective immunity by host mimicry. As HIV-1 nNAbs isolated from infected patients are not significantly enriched for poly- and autoreactivity (Fig. 5 and and6),6), these structural traits are unlikely the consequence of the HIV-1 infection milieu but rather linked to neutralization potency. This link is independently supported by the CH103 Ab lineage, where virus neutralizing breadth was associated with increasing polyreactivity (7). Given that bNAbs and nNAbs can recognize overlapping epitopes on HIV-1, why are bNAbs alone selected for polyreactivity? One explanation for bNAb polyreactivity is Ab heteroligation that increases effective binding affinity and neutralization capacity (23, 28). This model, however, is difficult to reconcile with infrequent poly- and autoreactivity among autologous neutralizing nNAbs (Fig. 5 and and6).6). In contrast, our hypothesis that conserved, HIV-1 neutralizing epitopes mimic host Ags is consistent with the disparate patterns of poly/autoreactivity between neutralizing bNAbs and nNAbs. nNAbs recognize evanescent, neutralizing epitopes that are readily altered in virus mutants but have little effect on viral fitness (58); bNAbs, on the other hand, bind epitopes crucial to viral fitness and resistant to change by mutation (59). It is unsurprising that these crucial epitopes might undergo intense selection favoring host mimicry and other forms of immune evasion. Host mimicry limits the effective repertoire of bNAb B cells to increase viral fitness (18, 24, 60); if the germ line ancestors of bNAb B cells are absent or rare in the primary B-cell repertoire, extraordinary levels of hypermutation may be the only pathway for peripheral B cells to acquire bNAb activity (61). As hypermutation in germinal center B cells generates de novo autoreactivity (62), we wonder whether the increased poly- and autoreactivity among bNAbs (Fig. 5 and and6)6) is necessary for neutralization (23, 28) or a by-product of tortuous clonal evolution (61).
Our survey of bNAb specificity may be clinically relevant. Passive bNAb immunization is being considered for acute HIV-1 infection (63) and to prevent maternal-fetal transmission of HIV-1 (64). Poly- or autoreactive bNAbs may reduce serum half-lives (65) or induce pathology. Finally, we note that induction of bNAbs may require immunization strategies that (transiently) bypass or suppress immunological tolerance (61).
ACKNOWLEDGMENTS
This work was supported by NIAID grants AI 81579 and AI 100645 and the Bill and Melinda Gates Foundation. M.L. was a Howard Hughes Medical Fellow.
We declare that we have no financial conflicts of interest.
We thank J. Zhang, X. Liang, and D. Liao for expert assistance and J. St. Geme, M. Kuraoka, D. Cain, and T. Nojima for advice. M. Connors, P. J. Bjorkman, and J. R. Mascola kindly provided the following antibodies: 10E8 bNAb (M. Connors), NIH45-46 bNAb (P. J. Bjorkman), and VRC01, VRC02, VRC03, and VRC07 bNAbs (J. R. Mascola).
G.K. and B.F.H. conceived the study. G.K. designed and supervised all experiments. M.L. and G.Y. designed and conducted experiments and analyzed data. K.W. and N.I.N. provided structural analyses. N.A.V. and W.R. designed and performed statistical analyses. S.M.A. performed and analyzed SPR. J.G. generated recombinant wild-type and mutant UBE3A. M.B. gave advice and reagents. G.Y., M.L., B.F.H., and G.K. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02378-14.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.02378-14
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/89/1/784.full.pdf
Citations & impact
Impact metrics
Article citations
Isolation and characterization of IgG3 glycan-targeting antibodies with exceptional cross-reactivity for diverse viral families.
PLoS Pathog, 20(9):e1012499, 18 Sep 2024
Cited by: 0 articles | PMID: 39292703 | PMCID: PMC11410209
Human antibody polyreactivity is governed primarily by the heavy-chain complementarity-determining regions.
Cell Rep, 43(10):114801, 10 Oct 2024
Cited by: 0 articles | PMID: 39392756 | PMCID: PMC11564698
Affinity gaps among B cells in germinal centers drive the selection of MPER precursors.
Nat Immunol, 25(6):1083-1096, 30 May 2024
Cited by: 2 articles | PMID: 38816616 | PMCID: PMC11147770
Broadly neutralizing antibodies for HIV prevention: a comprehensive review and future perspectives.
Clin Microbiol Rev, 37(2):e0015222, 30 Apr 2024
Cited by: 1 article | PMID: 38687039
Review
Inhibition of human immunodeficiency virus (HIV-1) infectivity by expression of poorly or broadly neutralizing antibodies against Env in virus-producing cells.
J Virol, 98(2):e0159423, 30 Jan 2024
Cited by: 0 articles | PMID: 38289101 | PMCID: PMC10878270
Go to all (128) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
RefSeq - NCBI Reference Sequence Database
- (1 citation) RefSeq - NM_000462.3
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Broadly Neutralizing Antibodies Display Potential for Prevention of HIV-1 Infection of Mucosal Tissue Superior to That of Nonneutralizing Antibodies.
J Virol, 91(1):e01762-16, 16 Dec 2016
Cited by: 24 articles | PMID: 27795431 | PMCID: PMC5165208
Lack of ADCC Breadth of Human Nonneutralizing Anti-HIV-1 Antibodies.
J Virol, 91(8):e02440-16, 29 Mar 2017
Cited by: 52 articles | PMID: 28122982 | PMCID: PMC5375671
HIV-1 Subtype C-Infected Children with Exceptional Neutralization Breadth Exhibit Polyclonal Responses Targeting Known Epitopes.
J Virol, 92(17):e00878-18, 16 Aug 2018
Cited by: 39 articles | PMID: 29950423 | PMCID: PMC6096808
Poly- and autoreactivity of HIV-1 bNAbs: implications for vaccine design.
Retrovirology, 15(1):53, 28 Jul 2018
Cited by: 14 articles | PMID: 30055635 | PMCID: PMC6064052
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIAID NIH HHS (4)
Grant ID: AI 100645
Grant ID: UM1 AI100645
Grant ID: AI 81579
Grant ID: R01 AI081579
 b,c
b,c




