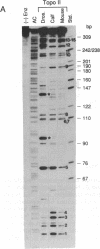
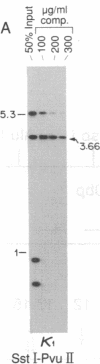
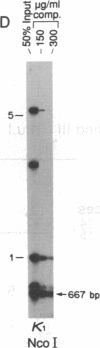
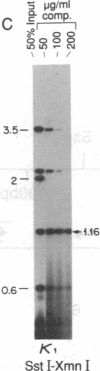
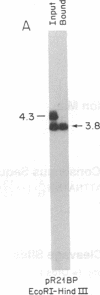
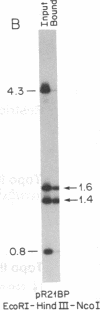
Abstract
Free full text

Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II.
Abstract
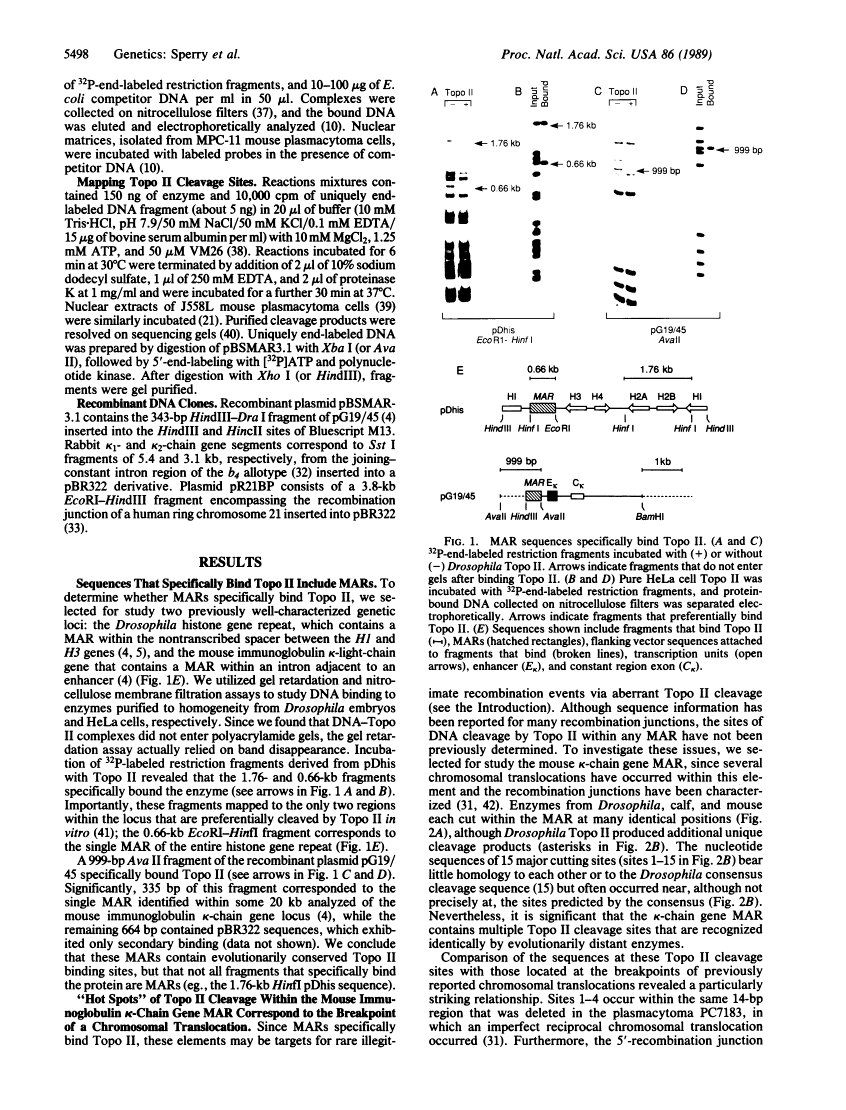
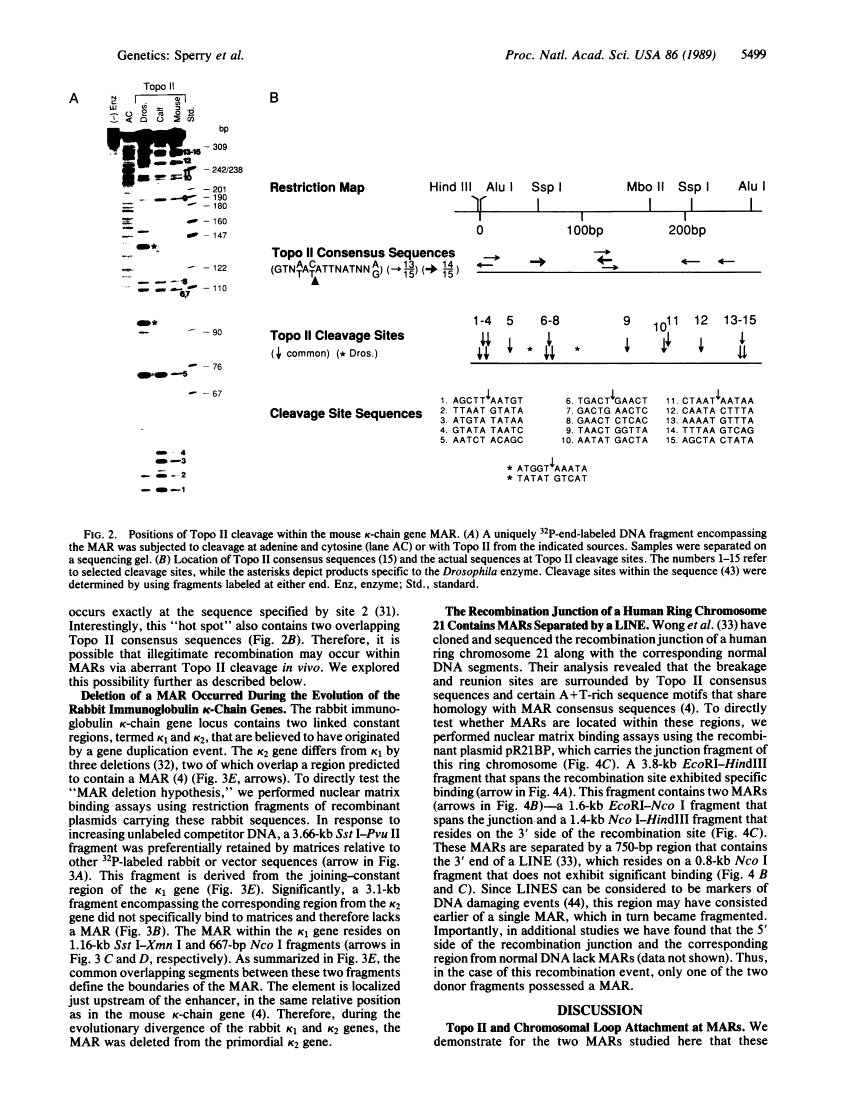
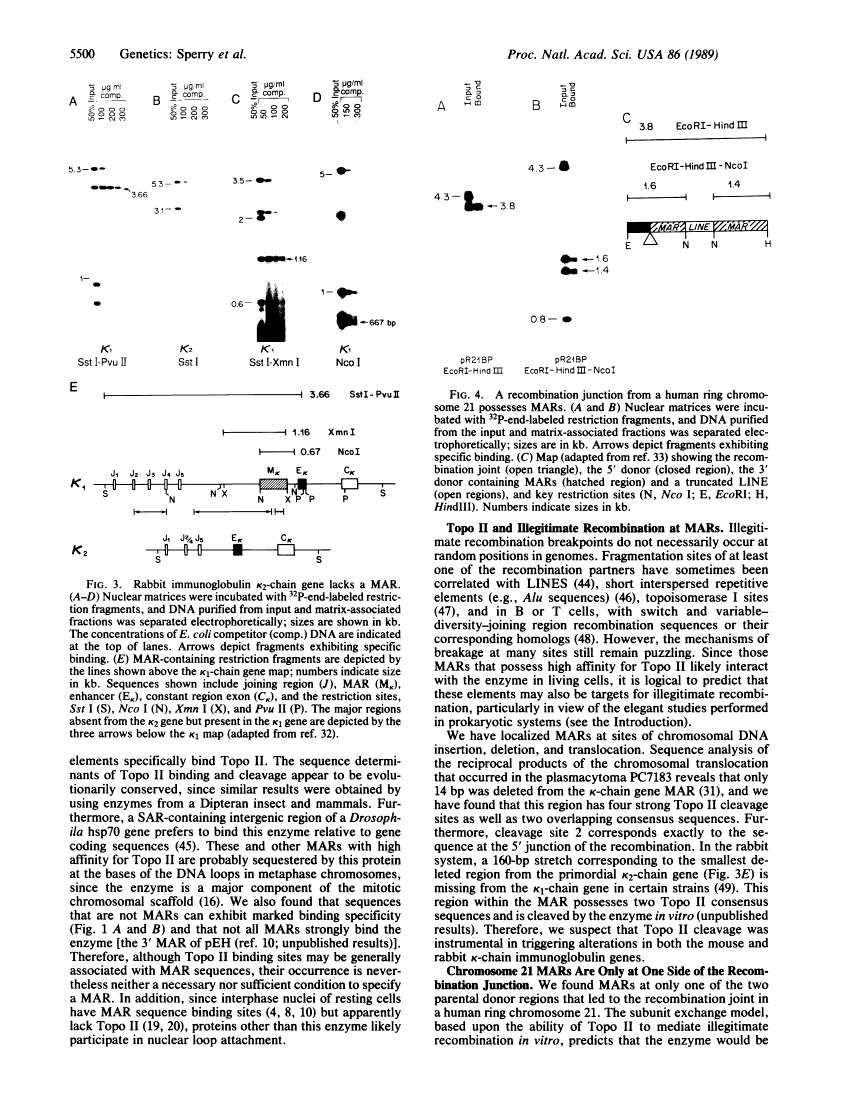
A family of A + T-rich sequences termed MARs ("matrix association regions") mediate chromosomal loop attachment. Here we demonstrate that several MARs both specifically bind and contain multiple sites of cleavage by topoisomerase II, a major protein of the mitotic chromosomal scaffold. Interestingly, "hotspots" of enzyme cutting occur within the MAR of the mouse immunoglobulin kappa-chain gene at the breakpoint of a previously described chromosomal translocation. Since topoisomerase II can mediate illegitimate recombination in prokaryotes, we explored further the possibility that MARs might be targets for this process in eukaryotes. We found that a MAR had been deleted from one of the two rabbit immunoglobulin kappa-chain genes and that MARs reside next to a long interspersed repetitive element within the recombination junction of a human ring chromosome 21. These results, taken together with other accounts of nonhomologous recombination, lead to the proposal that a dysfunction of MARs is illegitimate recombination.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Worcel A, Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. [Abstract] [Google Scholar]
- Benyajati C, Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. [Abstract] [Google Scholar]
- Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. [Abstract] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. [Abstract] [Google Scholar]
- Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. [Abstract] [Google Scholar]
- Gasser SM, Laemmli UK. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. [Abstract] [Google Scholar]
- Izaurralde E, Mirkovitch J, Laemmli UK. Interaction of DNA with nuclear scaffolds in vitro. J Mol Biol. 1988 Mar 5;200(1):111–125. [Abstract] [Google Scholar]
- Loc PV, Strätling WH. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. [Europe PMC free article] [Abstract] [Google Scholar]
- Cockerill PN, Garrard WT. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 1986 Aug 11;204(1):5–7. [Abstract] [Google Scholar]
- Cockerill PN, Yuen MH, Garrard WT. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987 Apr 15;262(11):5394–5397. [Abstract] [Google Scholar]
- Gasser SM, Laemmli UK. The organisation of chromatin loops: characterization of a scaffold attachment site. EMBO J. 1986 Mar;5(3):511–518. [Europe PMC free article] [Abstract] [Google Scholar]
- Amati BB, Gasser SM. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. [Abstract] [Google Scholar]
- Mirkovitch J, Spierer P, Laemmli UK. Genes and loops in 320,000 base-pairs of the Drosophila melanogaster chromosome. J Mol Biol. 1986 Jul 20;190(2):255–258. [Abstract] [Google Scholar]
- Mirkovitch J, Gasser SM, Laemmli UK. Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol. 1988 Mar 5;200(1):101–109. [Abstract] [Google Scholar]
- Sander M, Hsieh TS. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. [Europe PMC free article] [Abstract] [Google Scholar]
- Earnshaw WC, Heck MM. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985 May;100(5):1716–1725. [Europe PMC free article] [Abstract] [Google Scholar]
- Berrios M, Osheroff N, Fisher PA. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4142–4146. [Europe PMC free article] [Abstract] [Google Scholar]
- Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987 Jan 30;48(2):219–230. [Abstract] [Google Scholar]
- Heck MM, Earnshaw WC. Topoisomerase II: A specific marker for cell proliferation. J Cell Biol. 1986 Dec;103(6 Pt 2):2569–2581. [Europe PMC free article] [Abstract] [Google Scholar]
- Fairman R, Brutlag DL. Expression of the Drosophila type II topoisomerase is developmentally regulated. Biochemistry. 1988 Jan 26;27(2):560–565. [Abstract] [Google Scholar]
- Darby MK, Herrera RE, Vosberg HP, Nordheim A. DNA topoisomerase II cleaves at specific sites in the 5' flanking region of c-fos proto-oncogenes in vitro. EMBO J. 1986 Sep;5(9):2257–2265. [Europe PMC free article] [Abstract] [Google Scholar]
- Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. [Abstract] [Google Scholar]
- Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988 Dec 2;55(5):849–856. [Abstract] [Google Scholar]
- Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. [Abstract] [Google Scholar]
- Gaulden ME. Hypothesis: some mutagens directly alter specific chromosomal proteins (DNA topoisomerase II and peripheral proteins) to produce chromosome stickiness, which causes chromosome aberrations. Mutagenesis. 1987 Sep;2(5):357–365. [Abstract] [Google Scholar]
- Bae YS, Kawasaki I, Ikeda H, Liu LF. Illegitimate recombination mediated by calf thymus DNA topoisomerase II in vitro. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2076–2080. [Europe PMC free article] [Abstract] [Google Scholar]
- Ikeda H, Kawasaki I, Gellert M. Mechanism of illegitimate recombination: common sites for recombination and cleavage mediated by E. coli DNA gyrase. Mol Gen Genet. 1984;196(3):546–549. [Abstract] [Google Scholar]
- O'Connor MB, Malamy MH. Mapping of DNA gyrase cleavage sites in vivo oxolinic acid induced cleavages in plasmid pBR322. J Mol Biol. 1985 Feb 20;181(4):545–550. [Abstract] [Google Scholar]
- Ikeda H. Bacteriophage T4 DNA topoisomerase mediates illegitimate recombination in vitro. Proc Natl Acad Sci U S A. 1986 Feb;83(4):922–926. [Europe PMC free article] [Abstract] [Google Scholar]
- Ripley LS, Dubins JS, deBoer JG, DeMarini DM, Bogerd AM, Kreuzer KN. Hotspot sites for acridine-induced frameshift mutations in bacteriophage T4 correspond to sites of action of the T4 type II topoisomerase. J Mol Biol. 1988 Apr 20;200(4):665–680. [Abstract] [Google Scholar]
- Shapiro MA, Weigert M. A complex translocation at the murine kappa light-chain locus. Mol Cell Biol. 1987 Nov;7(11):4130–4133. [Europe PMC free article] [Abstract] [Google Scholar]
- Emorine L, Max EE. Structural analysis of a rabbit immunoglobulin kappa 2 J-C locus reveals multiple deletions. Nucleic Acids Res. 1983 Dec 20;11(24):8877–8890. [Europe PMC free article] [Abstract] [Google Scholar]
- Wong C, Kazazian HH, Jr, Stetten G, Earnshaw WC, Van Keuren ML, Antonarakis SE. Molecular mechanism in the formation of a human ring chromosome 21. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1914–1918. [Europe PMC free article] [Abstract] [Google Scholar]
- Shelton ER, Osheroff N, Brutlag DL. DNA topoisomerase II from Drosophila melanogaster. Purification and physical characterization. J Biol Chem. 1983 Aug 10;258(15):9530–9535. [Abstract] [Google Scholar]
- Miller KG, Liu LF, Englund PT. A homogeneous type II DNA topoisomerase from HeLa cell nuclei. J Biol Chem. 1981 Sep 10;256(17):9334–9339. [Abstract] [Google Scholar]
- Trask DK, DiDonato JA, Muller MT. Rapid detection and isolation of covalent DNA/protein complexes: application to topoisomerase I and II. EMBO J. 1984 Mar;3(3):671–676. [Europe PMC free article] [Abstract] [Google Scholar]
- Berent SL, Sevall JS. Histone H1 binding at the 5' end of the rat albumin gene. Biochemistry. 1984 Jun 19;23(13):2977–2983. [Abstract] [Google Scholar]
- Osheroff N, Zechiedrich EL. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987 Jul 14;26(14):4303–4309. [Abstract] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. [Europe PMC free article] [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. [Abstract] [Google Scholar]
- Udvardy A, Schedl P, Sander M, Hsieh TS. Novel partitioning of DNA cleavage sites for Drosophila topoisomerase II. Cell. 1985 Apr;40(4):933–941. [Abstract] [Google Scholar]
- Perlmutter RM, Klotz JL, Pravtcheva D, Ruddle F, Hood L. A novel 6:10 chromosomal translocation in the murine plasmacytoma NS-1. Nature. 1984 Feb 2;307(5950):473–476. [Abstract] [Google Scholar]
- Max EE, Maizel JV, Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [Abstract] [Google Scholar]
- Sander M, Hsieh T, Udvardy A, Schedl P. Sequence dependence of Drosophila topoisomerase II in plasmid relaxation and DNA binding. J Mol Biol. 1987 Mar 20;194(2):219–229. [Abstract] [Google Scholar]
- Lehrman MA, Schneider WJ, Südhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985 Jan 11;227(4683):140–146. [Europe PMC free article] [Abstract] [Google Scholar]
- Bullock P, Champoux JJ, Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. [Abstract] [Google Scholar]
- Croce CM. Role of chromosome translocations in human neoplasia. Cell. 1987 Apr 24;49(2):155–156. [Abstract] [Google Scholar]
- Akimenko MA, Mariamé B, Rougeon F. Evolution of the immunoglobulin kappa light chain locus in the rabbit: evidence for differential gene conversion events. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5180–5183. [Europe PMC free article] [Abstract] [Google Scholar]
- Orr-Weaver TL, Szostak JW. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. [Europe PMC free article] [Abstract] [Google Scholar]
- Anand R, Boehm CD, Kazazian HH, Jr, Vanin EF. Molecular characterization of a beta zero-thalassemia resulting from a 1.4 kilobase deletion. Blood. 1988 Aug;72(2):636–641. [Abstract] [Google Scholar]
- Jarman AP, Higgs DR. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. [Europe PMC free article] [Abstract] [Google Scholar]
- Dijkwel PA, Hamlin JL. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. [Europe PMC free article] [Abstract] [Google Scholar]
- Hyrien O, Debatisse M, Buttin G, de Saint Vincent BR. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J. 1987 Aug;6(8):2401–2408. [Europe PMC free article] [Abstract] [Google Scholar]
- Vanin EF, Henthorn PS, Kioussis D, Grosveld F, Smithies O. Unexpected relationships between four large deletions in the human beta-globin gene cluster. Cell. 1983 Dec;35(3 Pt 2):701–709. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.86.14.5497
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/86/14/5497.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Activation of recombinational repair in Ewing sarcoma cells carrying EWS-FLI1 fusion gene by chromosome translocation.
Sci Rep, 12(1):14764, 30 Aug 2022
Cited by: 1 article | PMID: 36042341 | PMCID: PMC9427769
Weak interactions in higher-order chromatin organization.
Nucleic Acids Res, 48(9):4614-4626, 01 May 2020
Cited by: 29 articles | PMID: 32313950 | PMCID: PMC7229822
Review Free full text in Europe PMC
Matrix association region/scaffold attachment region: the crucial player in defining the positions of chromosome breaks mediated by bile acid-induced apoptosis in nasopharyngeal epithelial cells.
BMC Med Genomics, 12(1):9, 15 Jan 2019
Cited by: 0 articles | PMID: 30646906 | PMCID: PMC6334432
Are TADs supercoiled?
Nucleic Acids Res, 47(2):521-532, 01 Jan 2019
Cited by: 30 articles | PMID: 30395328 | PMCID: PMC6344874
Review Free full text in Europe PMC
MAR-Mediated transgene integration into permissive chromatin and increased expression by recombination pathway engineering.
Biotechnol Bioeng, 114(2):384-396, 03 Oct 2016
Cited by: 16 articles | PMID: 27575535 | PMCID: PMC5215416
Go to all (98) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Protein:DNA interactions at chromosomal loop attachment sites.
Genome, 31(2):503-509, 01 Jan 1989
Cited by: 41 articles | PMID: 2561108
Review
The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements.
J Biol Chem, 262(11):5394-5397, 01 Apr 1987
Cited by: 128 articles | PMID: 3031052
Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites.
Cell, 44(2):273-282, 01 Jan 1986
Cited by: 537 articles | PMID: 3002631
Illegitimate recombination mediated by calf thymus DNA topoisomerase II in vitro.
Proc Natl Acad Sci U S A, 85(7):2076-2080, 01 Apr 1988
Cited by: 82 articles | PMID: 2832845 | PMCID: PMC279931
Funding
Funders who supported this work.
NIGMS NIH HHS (3)
Grant ID: GM22201
Grant ID: GM29935
Grant ID: GM31689