Abstract
Free full text

A genome-wide association study reveals the quantitative trait locus and candidate genes that regulate phosphate efficiency in a Vietnamese rice collection
Associated Data
Abstract
The crucial role of phosphate (Pi) for plant alongside the expected depletion of non-renewable phosphate rock have created an urgent need for phosphate-efficient rice varieties. In this study, 157 greenhouse-grown Vietnamese rice landraces were treated under Pi-deficient conditions to discover the genotypic variation among biochemical traits, including relative efficiency of phosphorus use (REP), relative root to shoot weight ratio (RRSR), relative physiological phosphate use efficiency (RPPUE), and relative phosphate uptake efficiency (RPUpE). Plants were grown in Yoshida nutrient media with either a full (320 μM) or a low Pi supply (10 μM) over six weeks. This genome-wide association study led to the discovery of 31 significant single nucleotide polymorphisms, 18 quantitative trait loci (QTLs), and 85 candidate genes. A common QTL named qRPUUE9.16 was found among the three investigated traits. Some interesting candidate genes, such as PLASMA MEMBRANE PROTEIN1 (OsPM1), CALMODULIN-RELATED CALCIUM SENSOR PROTEIN 15 (OsCML15), phosphatases 2C (PP2C), STRESS-ACTIVATED PROTEIN KINASE (OsSAPK2), and GLYCEROPHOSPHORYL DIESTER PHOSPHODIESTERASES (GDPD13), were found strongly correlated to the Pi starvation. RNA sequencing transcriptomes revealed that 45 out of 85 candidate genes were significantly regulated under Pi starvation. Furthermore, nearly two-thirds of genotypes did not possess the OsPsTOL1 gene; however, no significant difference was observed in response to Pi deficiency between genotypes with or without this gene, suggesting that other QTLs in rice may resist Pi starvation. These results provide new information on the genetics of nutrient use efficiency in rice and may potentially assist with developing more phosphate-efficient rice plants.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00902-2) contains supplementary material, which is available to authorized users.
Introduction
Fertilization has been utilized as the chief mechanism to improve crop productivity since the launch of green revolution’s initiatives. Essential mineral elements, such as nitrogen, phosphorus (P), and potassium which can be found in fertilizers, have drawn special attention from scientists. Phosphorus, which ranks second in the macro-element group, is a component of sugar-phosphate, nucleic acids, and phospholipids (Taiz and Zeiger 2002); thus, has an imperative role in photosynthesis as well as the growth and development of plants. Deficiency of inorganic phosphate (Pi), the form of P which is accessible to plants, in cereal crops can severely affect plant productivity (Raghothama 1999; Balemi and Negisho 2012). Specifically, rice often displays stunted shoot development mainly owing to low Pi in medium, which consequently leads to poor grain quality and low yield (Kennelly et al. 2012). Unfortunately, the usable Pi content in arable soils tends to be deficient, although the total amount of Pi in soil may still be adequate (Vance et al. 2003). This is because Pi often forms insoluble complexes with iron and aluminum in acidic conditions (Lindsay 1979) or with calcium in alkaline conditions (Haefele et al. 2014), which leads to Pi starvation in approximately 70% of globally cultivated land (Kirkby and Johnston 2008). In addition, the use of non-renewable Pi rock-derived fertilizers not only further diminishes resources (Gilbert 2009) but also poses ecological risks to adjacent water bodies and natural habitats (Le et al. 2010). Therefore, the long-term solutions to meet the needs of worldwide food security while also avoiding the negative impacts of insufficient Pi certainly require improvements in plant phosphate acquisition efficiency (PAE), phosphate uptake efficiency (PUpE) and phosphate use efficiency (PUE) (Lizbeth et al. 2014; MacDonald et al. 2011; Stigter and Plaxton 2015).
Different genetic engineering approaches such as activation of Pi transporters, Pi-regulatory proteins, purple acid phosphatase, and the proteins responsible for organic acid synthesis as well as modification of root architecture have been developed to enhance PUpE, PAE, and PUE. Indeed, several studies have demonstrated the role of Pi transporters in mediating Pi uptake at the root and soil interface to overcome Pi starvation in plants (Mudge et al. 2002; Schünmann et al. 2004). Under Pi starvation, expression levels of the Pi transporter (Pht) genes OsPht1;8 and OsPht1;1 in rice (Oryza sativa) (Sun et al. 2012; Li et al. 2015); Pht1;1, Pht1;2, Pht1;3, and Pht1;4 in Arabidopsis thaliana (Mudge et al. 2002); HvPht1;1 in barley (Hordeum vulgare L.) (Schünmann et al. 2004); and TaPht1.2 and TaPht1.4 in wheat (Triticum aestivum) (Miao et al. 2009; Liu et al. 2013) have been observed as increasing significantly. The overexpression of Pi transporters has been extensively addressed in diverse plant species. For instance, in A. thaliana, the notable production of Pi in siliques and increase in the expression levels of genes involved in Pi scavenging resulted from the overexpression of AtPht1;5 (Nagarajan et al. 2011).
Furthermore, in OsPht1;1-overexpressed rice plants, a two-fold increase in shoot Pi content as well as higher numbers of tillers and panicles were observed relative to wild-type plants (Seo et al. 2008). Overexpression of OsPHT1;6 also resulted in higher Pi content and a higher grain yield in transgenic rice plants (Zhang et al. 2014). In addition, manipulating transcription factors to improve PUpE, PAE, and PUE has been considered a promising approach. Overexpressing PHOSPHATE STARVATION RESPONSE 1 (OsPHR1) in A. thaliana has been reported to improve Pi uptake (Nilsson et al. 2007). The same phenomenon was observed when TaPHR1-A1, an ortholog of OsPHR1, was overexpressed in T. aestivum (Wang et al. 2013).
Modification of root architecture may be a crucial strategy for responding to Pi starvation. A number of quantitative trait loci (QTLs) associated with root traits and PUE have thus been identified. For example, a low Pi tolerance QTL named phosphate uptake 1 (Pup1) was discovered in the traditional rice accession Kasalath (Wissuwa et al. 1998; Wissuwa and Ae 2001). Then, the PHOSPHORUS STARVATION TOLERANCE 1 (OsPSTOL1) gene, which encodes for a serine/threonine kinase, was identified within the Pup1 region in the Kasalath rice accession. Overexpression of this gene in the Nipponbare and IR64 rice accessions led to increase of up to 60% higher grain yield compared to wild-type plants under Pi starvation (Gamuyao et al. 2012). Therefore, OsPSTOL1 has been considered as a major genetic determinant of low Pi tolerance in rice.
However, in another study conducted by Yumnam et al. (2017) the nucleotide polymorphisms in OsPSTOL1 and PupK20-2, which were reported for low Pi tolerance, were evaluated. The results showed two distinct haplotypes in the OsPSTOL1 sequence in which two out of five tolerant genotypes had the exact Kasalath-type haplotype, whereas others showed a mixed haplotype. In the 3′-UTR (untranslated region) of OsPSTOL1, four new nucleotide variations were observed, and 28 single nucleotide polymorphisms (SNPs) as well as one insertion–deletion mutation were identified in OsPupK20-2 (Yumnam et al. 2017). Furthermore, a novel OsPSTOL1 allele containing 35 base pair substitution compared to the Kasalath allele was detected in upland New Rice for Africa (NERICA) varieties and most O. glaberrima accessions in Africa (Pariasca-Tanaka et al. 2014). In addition, a G to A nucleotide substitution that led to an early stop mutation and a 2 base pair frameshift deletion was observed in a collection of 282 rice accessions (Vigueira et al. 2016). These results strongly suggest the presence of other tolerant gene(s) in these genotypes.
Subsequently, another QTL named root elongation under phosphorus deficiency 6 (qREP-6) was found to demonstrate a positive correlation between root elongation and Pi content in shoots and between root elongation and tiller number under Pi starvation conditions (Shimizu et al. 2008). In addition, researchers also identified several other QTLs, including qMRL6a (maximum root length), qRN8b (root number), and qRN4 (root number) under Pi starvation (Li et al. 2009). Aside from QTLs related to root traits, several other QTLs related to total above-ground Pi uptake (qPUP1, qPUP7, and qPUP10), Pi harvest index (qPHI1, qPHI2, qPHI6, and qPHI11), PUE for grain yield (qgPUE4), PUE for straw dry weight (qstrPUE1-1, qstrPUE1-2, and qstrPUE2), and PUE for biomass accumulation (qPUEb2) were identified by Wang and his colleagues (Wang et al. 2014).
Genome-wide association study (GWAS) is a powerful tool for identifying the associations between SNPs and traits in a large population. GWAS assesses if genetic variants are associated with a specific trait (Manolio 2010). In rice, GWAS has been applied to study various traits related to metabolites (Chen et al. 2014), leaf traits (Yang et al. 2015), and salinity stress (Lekklar et al. 2019). However, studies on rice tolerance to Pi starvation using a large variety of rice collections remain conspicuously absent. In 2015, Wissuwa et al. (2015) used 282 rice accessions to study internal PUE using a GWAS approach. As a result, six loci associated with PUE were detected. Under Pi starvation conditions, two loci were found to be linked to root biomass production, whereas the remaining four were either specific to shoots or total biomass (Wissuwa et al. 2015). Thus, four QTLs of interest were identified; however, only one QTL located on chromosome 1 was chosen for further studies as a minor haplotype increasing PUE, which could be applied to breeding programs. Among 14 annotated genes on chromosome 1 (7.22–7.34 Mb), PUE1-7 was identified as the most responsive gene to Pi availability. In response to Pi starvation, this gene was significantly upregulated, especially in old leaves, suggesting its function in the remobilization of Pi. PUE1-12 and PUE1-13 in the PUE1 region may also be further considered as they were differentially expressed under Pi starvation (Wissuwa et al. 2015).
Furthermore, a Vietnamese genotyping study involving the sequencing of 21,971 SNP markers from 182 Vietnamese rice varieties has helped to improve our understanding of rice diversity and support further GWAS analyses (Phung et al. 2014). Therefore, in this study, 157 Vietnamese rice accessions from the aforementioned collection were used to study the response of diverse local rice varieties to Pi deficient conditions using GWAS analysis. Four biochemical traits, including relative root to shoot ratio (RRSR), relative efficiency of P use (REP), relative physiological PUE (RPPUE), and relative PUpE (RPUpE) were assessed. In addition, a list of candidate genes in proximity to the most significant markers were identified to extract novel information on the adaptability of rice to low Pi conditions.
Materials and methods
Plant materials and genotyping data
A population of 157 rice landraces originating from diverse provinces and ecosystems (irrigated, rainfed lowland, mangrove, or upland) with different types (traditional or improved) was selected from the 182 characterized varieties in the public panel of Vietnamese rice accessions (Phung et al. 2014). Their seeds were provided by the Plant Resources Center in Hanoi, Vietnam. The list of rice accessions and their related information are provided in Supplementary Table 1. A total of 25,971 SNP markers, which were previously identified by genotype sequencing (Phung et al. 2014), were used in this study.
Plant growth
The seeds were first incubated in an oven at 45 °C for 5 days to break down seed dormancy. Then, the seeds were germinated at 37 °C over 3–4 days. The plantlets were grown in controlled greenhouse conditions at 28–30 °C and approximately 70–80% humidity. To avoid the block effect, the plants were randomly positioned using IRRISTAT software v4.0 (International Rice Research Institute (IRRI), Los Baños, Philippines) with three replicates. Individual plantlet was grown in drainable black plastic bags measuring 68 cm ×
× 16 cm in length and diameter and filled with fine river sand. The plants were grown in two different Yoshida nutrient conditions (Yoshida et al. 1971): one received a full Pi supplement (P0) of 320 μM, whereas the other received a low Pi supplement (P*) of 10 μM. Specifically, 0.05 g/L NaH2PO4.H2O (320 μM P) was used in the full Pi medium, whereas 1.56 mg/L NaH2PO4.H2O (10 μM P) was used to supplement in the low Pi medium. The plants were irrigated with full Pi medium (control) or with the low Pi medium (treatment) every three days for six weeks.
16 cm in length and diameter and filled with fine river sand. The plants were grown in two different Yoshida nutrient conditions (Yoshida et al. 1971): one received a full Pi supplement (P0) of 320 μM, whereas the other received a low Pi supplement (P*) of 10 μM. Specifically, 0.05 g/L NaH2PO4.H2O (320 μM P) was used in the full Pi medium, whereas 1.56 mg/L NaH2PO4.H2O (10 μM P) was used to supplement in the low Pi medium. The plants were irrigated with full Pi medium (control) or with the low Pi medium (treatment) every three days for six weeks.
Sample harvesting and phenotyping experiment
The plants were harvested after six weeks. Any sand clinging to the plantlets’ root systems was carefully removed using light faucet water pressure. The plant samples were dried in an oven at 70 °C for up to one week until they reached a constant weight. Shoots and leaves, including the stem base, were weighed to obtain shoot weight (SHW), and a similar procedure was performed on the roots to obtain root weight (RTW).
The relative efficiency of Pi use (REP, %) was determined by dividing the plant dry mass (DM, g) under low Pi (10 µM) by the DM under high Pi (320 µM) according to Ozturk et al. (Ozturk et al. 2005).
The relative RTW/SHW ratio (RRSR) was obtained from the fraction of the RTW/SHW ratio under low Pi (10 µM) over the RTW/SHW ratio under high Pi (320 µM) (Ozturk et al. 2005).
The Pi content in rice shoots was analyzed using the vanadomolybdophosphoric acid colorimetric method (Rice et al. 2017). A standard curve was established with the corresponding vanadate–molybdate reagent and a range of Pi concentrations represented by hydrous KH2PO4. For each variety, 0.3 g of the homogeneous dry sample was subjected to dry ashing over a 6 h combustion process in a Muffle furnace (Nabertherm, New Castle, DE, USA). Wet ashing of the combusted sample by the acid hydrolysis method, which consisted of diluted HCl fuming (37%; Merck Millipore, Burlington, MA, USA) and concentrated HNO3 (69%; Merck Millipore) was subsequently performed. Absorbance of the assay solution was read at 420 nm by a UV-1800 UV–VIS spectrophotometer (Shimadzu, Kyoto, Japan). From the obtained optical density value, the Pi concentration in each sample was determined using the standard curve with its linear regression line.
The Pi content in shoots (mg) was calculated from the multiplication of Pi concentration (mg/g) and SHW (g). To obtain the PUpE (mg Pi/g Pi) of each Pi environment, the corresponding shoot Pi content was divided by sufficient or insufficient Pi supplies (Neto et al. 2016). The relative PUpE (RPUpE) was deduced as the ratio between the PUpE under low Pi and that under high Pi.
The PPUE (g2 SHW/mg Pi) was calculated as the fraction between the SHW and Pi concentration at a given Pi concentration in the rooting medium (Hammond et al. 2009). The relative PPUE (RPPUE) was derived by dividing the PPUE under low Pi by the PPUE under high Pi.
Presence and/or absence of OsPSTOL1 in rice collections
Genomic DNA from the leaves of 157 rice landraces was extracted using the cetyltrimethylammonium bromide method (Murray and Thompson 1980). A polymerase chain reaction (PCR) was performed to check for the presence of OsPSTOL1 in the Vietnamese rice collection. The sequence of the forward and reverse primers used in this study were 5′-ATGCTGCTCTGTCAAAGGGCAT-3′ and 5′-CAAGCTCAAAGCCCTTTTGGTG-3′, respectively (Mukherjee et al. 2014). The 20 µL reaction mix included 2 µL Dreamtaq buffer (10X) (Dreamtaq bufer; Thermo Fisher Scientific, Waltham, MA, USA), 0.75 units of DreamTaq DNA polymerase (Dreamtaq DNA polymerase; Thermo Fisher Scientific, Waltham, MA, USA), 200 nM deoxyribonucleotide triphosphates (dNTPs; Thermo Fisher Scientific, Waltham, MA, USA), 250 nM forward primer, 250 nM reverse primer, 100 ng DNA template, and H2O up to 20 µL. The following thermo cycle conditions were used: 95 °C for 4 min, 35 cycles at 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min. The reaction was terminated at 72 °C for 7 min. The PCR products were analyzed by 1% agarose gel electrophoresis at 100 V over 30 min.
Genome-wide association study
The correlation between the four investigated traits and the corresponding SNP loci was studied using the mixed linear model (MLM) implemented in TASSEL software v5.2.55. The structure matrix was identified using principal component analysis with six maximum alleles. The suggestive threshold of p <
< 3.0E-4 was employed to determine significance (To et al. 2019).
3.0E-4 was employed to determine significance (To et al. 2019).
Linkage disequilibrium (LD) heatmap and haplotype analysis
LD between SNPs was measured in terms of
Screening for candidate genes
To identify the potential candidate genes responsible for Pi starvation tolerance, the positions of significantly associated SNPs, which were clustered into previously identified QTLs, were used. The Michigan State University (MSU) Rice Genome Annotation Project Database Release 7 (https://rice.plantbiology.msu.edu) (Kawahara et al. 2013) was used to browse the expressed genes situated 25 kb upward and downward from each selected significant marker. A list of candidate genes with their annotated functions was generated.
In addition, the expression profiles of candidate genes related to low Pi treatment were also verified based on transcriptome data from a public data source (https://genevestigator.com) (Hruz et al. 2008). Only the genes expressed with changes greater than a definite value of 2 were selected.
Statistical analysis
The difference between different parameters was statistically analyzed using a one-way analysis of variance and the Student’s t-test in R software v3.6.
Results
Diversity in phenotypic responses to Pi deficiency in 157 rice accessions
In this study, four biochemical traits related to the Pi content in rice, including REP, RRSR, RPPUE, and RPUpE, were used to assess the diverse responses of 157 rice accessions to low Pi treatment (Fig. 1). The very low response of the REP trait showed that under low Pi conditions, rice plants strongly reduced their growth. Approximately, 96% of accessions had reduced REP values in low Pi conditions compared to a full Pi supply. Following the same trend, approximately one-third and one-half of accessions exhibited lower RRSR and RPPUE values in Pi starvation conditions compared to full Pi ones, respectively. However, in response to low Pi, a large increase in the RPUpE trait was recorded in all rice accession. An increase of up to 45-folds of RPUpE was observed in G3 and G131 plants grown under low Pi conditions compared to those grown with a full Pi supply (Fig. 1d). This phenomenon demonstrated that under Pi stress conditions, plants have mechanisms for the increased uptake of Pi for survival and development.
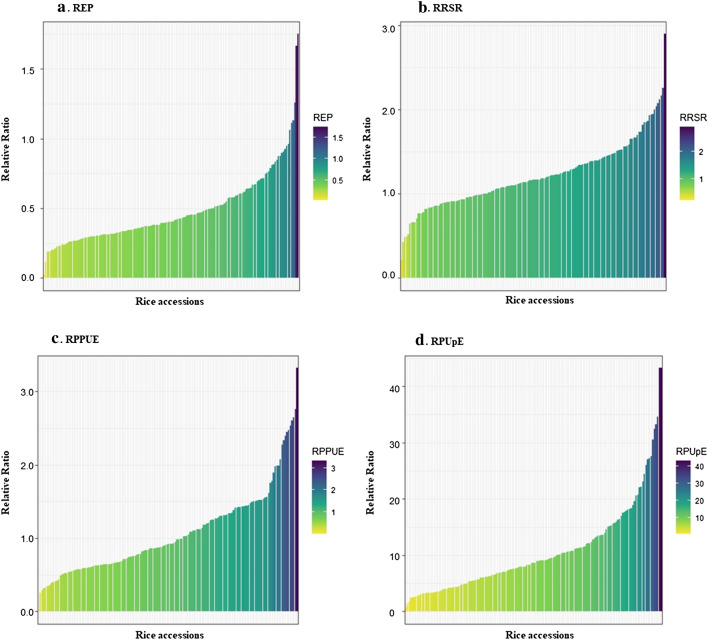
Phenotypic response of rice accessions to Pi starvation. Data was presented by relative values of a REP, b RSR, c PPUE and d PUpE traits under low and full Pi conditions
The phenotypic heterogeneity among the O. sativa subsp. indica and japonica was assessed and shown in Fig. 2. A significant difference in response to low Pi treatment between the indica and the japonica subgroups was recorded in the REP, RRSR, and RPPUE traits (Fig. 2a, 2b, and 2c), whereas no significant difference was found in the RPUpE trait (Fig. 2d). Moreover, the japonica subgroup had a lower rate of inhibition by low Pi treatment than indica as demonstrated by three significantly different REP, RRSR, and RPPUE traits. These data showed that subgroups of different genetic backgrounds respond differently to low Pi conditions.
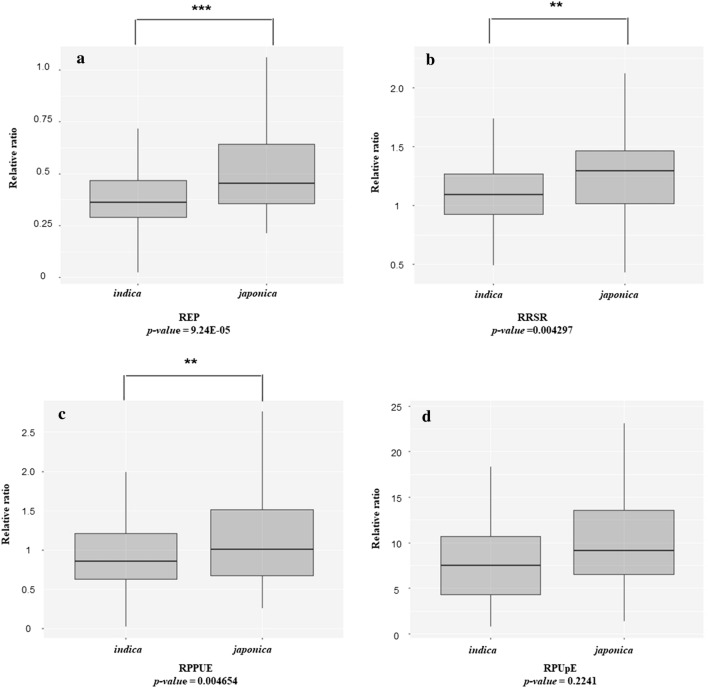
Phenotypic heterogeneity among indica and japonica sub-populations under Pi starvation conditions. Data was presented by values of a REP, b RRSR, c RPPUE and d RPUpE traits. The differences between sub-populations of each trait were analyzed using the Student's t-test. Asterisks (**), (***) indicates the significant difference with p value <
< 0.01 and 0.001, respectively
0.01 and 0.001, respectively
However, the response diversity was not observed in irrigated and upland ecosystems where only the RRSR trait displayed a significant difference in response to Pi starvation (Supplementary Fig. 3b). These ecosystems did not show any varying responses to low Pi conditions in the three other tested traits, including REP, RPPUE, and RPUpE (Supplementary Fig. 3a, 3c, and 3d).
PSTOL1-containing varieties
Sixty of the 157 rice accessions (38% of the collection) possessed the OsPsTOL1 gene (Supplementary Table 2). In this study, the correlation between the presence or absence of OsPsTOL1 and Pi-related traits was analyzed and shown in Fig. 3. Three out of four investigated traits, including RRSR, RPPUE, and RPUpE, showed no significant difference between accessions with OsPsTOL1 and those without OsPsTOL1 in response to low Pi treatment. In contrast, significantly higher REP in Pi-deficient soil conditions were observed in varieties not bearing OsPsTOL1 compared to those bearing this gene (Fig. 3a). Furthermore, many accessions, such as G3, G299, G62, and G131 were able to develop well under low-Pi conditions despite the absence of OsPsTOL1 in their genome. This data suggests the existence of another QTL beside Pup1 that can overcome low Pi conditions in the growth stage of our rice collection.
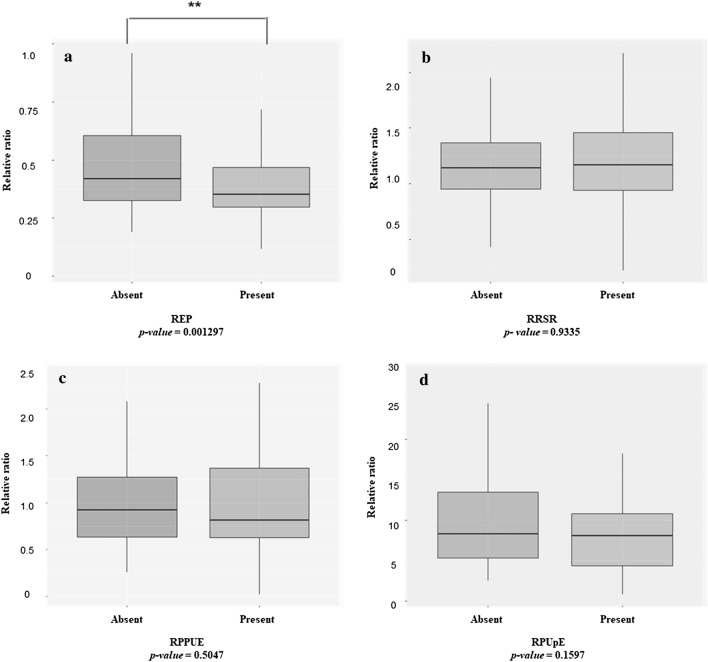
The correlation between with OsPSTOL1 and without OsPSTOL1-bearing varieties in a REP, b RRSR, c RPPUE and d RPUpE traits. The differences of REP, RRSR, RPPUE and RPUpE traits in the two types of rice accessions were analyszd using the Student's t-test. Asterisk (**) indicates the significant difference with p value <
< 0.01
0.01
Genome-wide association mapping
A GWAS was performed to identify the association between SNPs and four tested traits in the full rice panel using TASSEL software v.5. The threshold of p <
< 3.0E-4 was set as the significance level (To et al. 2019). The association results of the four traits were graphically displayed in Manhattan and Q-Q plots (Fig. 4). Overall, the Q-Q plot results showed fitted models between the experimental results and theoretical values. A total of 31 SNP-trait associations with p
3.0E-4 was set as the significance level (To et al. 2019). The association results of the four traits were graphically displayed in Manhattan and Q-Q plots (Fig. 4). Overall, the Q-Q plot results showed fitted models between the experimental results and theoretical values. A total of 31 SNP-trait associations with p <
< 3.0E-4 were detected. In detail, we identified seven common SNPs for REP, RPPUE, and RPUpE; two SNPs for only REP; two SNPs for only RRSR; 10 SNPs for only RPPUE; and 10 SNPs for only RPUpE. It should be noted that most of the significant SNPs residing on chromosomes 1 and 9 accounted for more than half of the total SNPs found in this study. The remaining SNPs were located on chromosomes 3, 4, 5, 7, 10, and 12. Detailed information regarding the positions and p-values of these significant markers can be found in Supplementary Table 3.
3.0E-4 were detected. In detail, we identified seven common SNPs for REP, RPPUE, and RPUpE; two SNPs for only REP; two SNPs for only RRSR; 10 SNPs for only RPPUE; and 10 SNPs for only RPUpE. It should be noted that most of the significant SNPs residing on chromosomes 1 and 9 accounted for more than half of the total SNPs found in this study. The remaining SNPs were located on chromosomes 3, 4, 5, 7, 10, and 12. Detailed information regarding the positions and p-values of these significant markers can be found in Supplementary Table 3.
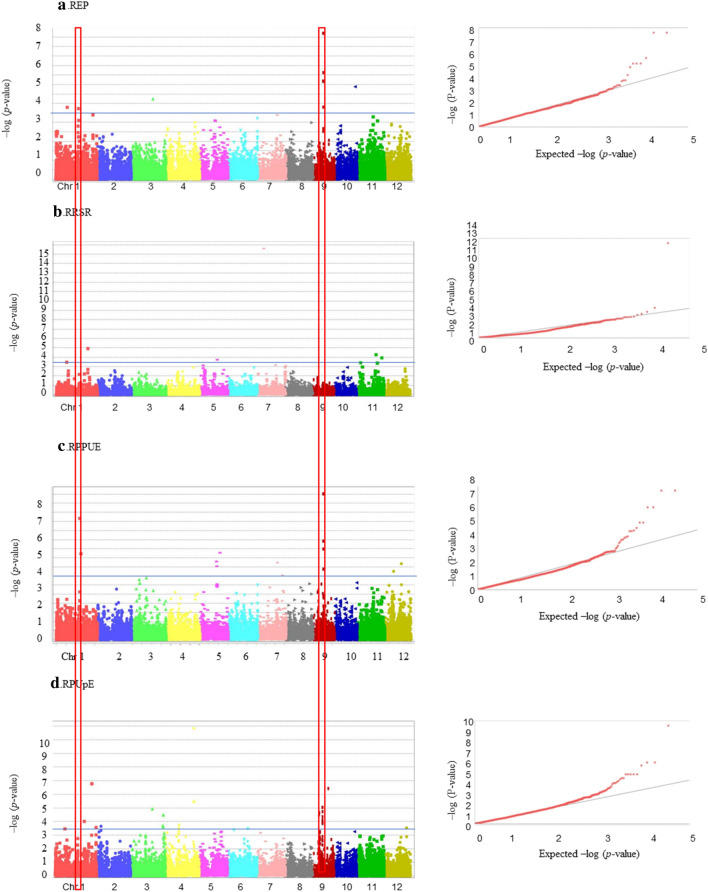
GWAS analysis the effect of Pi starvation on four investigated traits in the rice whole panel. Manhattan plot (left) and Q-Q plot (right) for a REP, b RRSR, c RPPUE and d RPUpE traits. The x-axis indicates the positions of SNPs across 12 chromosomes. The y-axis shows the log scale of p-value for the association test at each locus. The black line represents the significant threshold
QTL identification
Out of the 31 significant SNPs, 18 significant associations, including seven QTLs for RPPUE, two QTLs for RRSR, six QTLs for RPUpE, one common QTL for REP/RPPUE/RPUpE, and two QTLs for REP, were found from our GWAS analysis (Supplementary Table 3). The detected QTLs were distributed across eight chromosomes: 1, 3, 4, 5, 7, 9, 10, and 12. The significant SNPs in each QTL were used to assemble a haplotype sequence, followed by a polymorphism combination analysis was dissected for qRPUUE9.16 (stands for qREP9.16, qRPPUE9.16, and qRPUpE9.16), which was presented in three traits: RPPUE (Fig. 5b), REP (Fig. 5c), and RPUpE (Fig. 5d). Out of the 18 identified QTLs, qRPUUE9.16 was the largest at 57 kb. Moreover, according to Fig. 5a, the LD heatmap showed seven tightly linked markers in the qRPUUE9.16 QTL. A block of these seven closely associated markers indicated a strong linkage between them. Two main haplotypes, TTTTTTT and AAAAAAA, were used to compare the effect of haplotype on traits. The results showed that the accessions containing the AAAAAAA haplotype were significantly more affected than the ones containing the TTTTTTT haplotype under Pi starvation conditions, resulting in lower RPPUE and REP. Four candidate genes located on QTL qRPUUE9.16, including OsWAK86, OsLecRLK, OsGDPD13, and OsFBX318, were found (Fig. 5e).
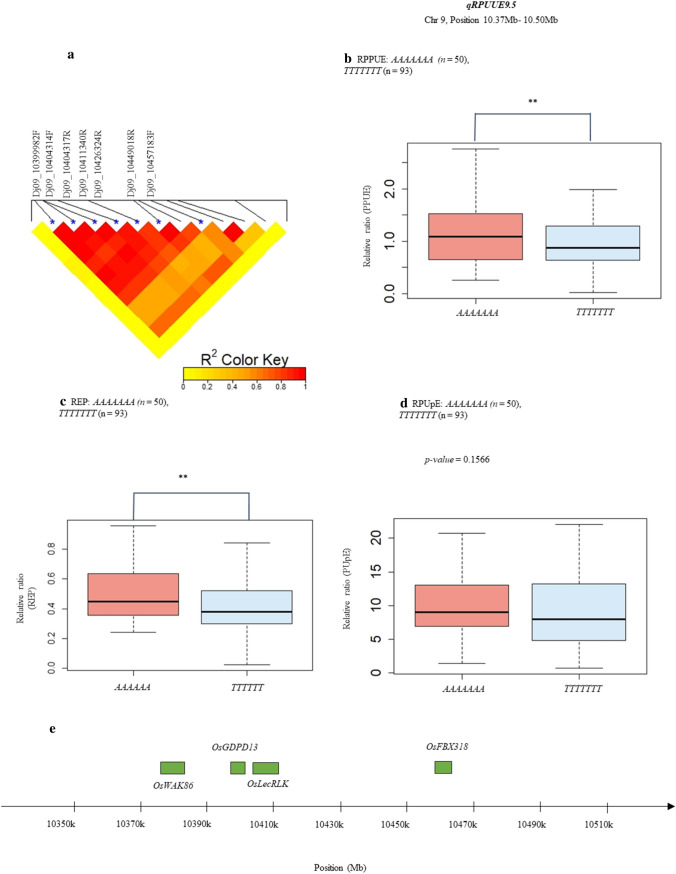
Haplotype analysis for the QTL qRPUUE9.16 on chromosome 9. a LD heatmaps in the peak region of association analysis GWAS for qRPUUE9.16. The asterisk (*) indicates significant SNPs. “Physical Length” indicates the total length of genetic region. b, c, d Effect of allelic combination of two main haplotypes on RPPUE, REP, RPUpE traits. N indicates the number of accessions for haplotype “AAAAAAA” and “TTTTTTT”. e Candidate genes and their positions on qRPUUE9.16. The level of significant difference between two haplotypes was analysed using the Student's t-test. Asterisk (**) indicates the significant difference with p-value <
< 0.01
0.01
Candidate gene localization in the proximity of significant SNPs derived from the GWAS results
Data from the Rice Genome Browser platform of the MSU Rice Genome Annotation Project Database Release 7 (https://rice.plantbiology.msu.edu) (Kawahara et al. 2013) was used to determine the candidate genes corresponding to each identified QTL of each trait in our study. The genes were identified within the 25 kb interval upward and downward of the most significant SNP derived from the GWAS results. A total of 85 expressed candidate genes were found to be located on chromosomes 1, 3, 4, 5, 7, 9, 10, and 12, and 27/85 and 10/85 genes were found on chromosomes 1 and 9, respectively (Supplementary Table 3). The Plant Transcription Factor Database (PlantTFDB) was used to search for transcription factors (TFs) (Wang et al. 2014). Out of the 85 expressed candidate genes, five TFs were found, including one bHLH (LOC_Os03g59670.1), two NACs (LOC_Os03g59730.1 and LOC_Os12g29330.1), one MYB (LOC_Os04g50680.1), and one G2-like (LOC_Os10g39550.1).
In addition, the RNA-Seq transcriptomes from a public database (https://genevestigator.com) (Hruz et al. 2008) were used to identify the expression profile of the candidate genes derived from our GWAS analysis relating to Pi treatment. Forty-five of the 85 candidate genes were regulated via an increase or decrease in expression levels under low Pi treatment with changes greater than a definite value of 2. Among these 45 genes, 14 genes were only upregulated, 10 genes were only downregulated, and 21 genes were both up—and downregulated depending on specific stress conditions (Supplementary Table 4). Among the five identified TFs, bHLH (LOC_Os03g59670.1) and NAC (LOC_Os12g29330.1) were found to be significantly regulated by low Pi treatment. Transcriptome information related to three other TFs has not been determined. Interestingly, in qRRSR5.11, LOC_Os05g31670.1, which encodes an AWPM-19-like membrane family protein, was highly upregulated up to 214.41 times more in the roots of the Nipponbare rice variety in treated medium without iron compared to medium without both iron and P. However, in the same QTL, LOC_Os05g31620.1, which encodes an OsCML15-calmodulin-related calcium sensor protein, was down-regulated 15.73 times more in the roots of a high tolerance to low Pi rice variety (Dular) compared to a more sensitive one (Pusa Basmati 1) in low Pi culture medium.
Discussion
In this study, the GWAS approach was carried out in a Vietnamese rice collection to identify QTLs associated with REP, RRSR, RPPUE, and RPUpE, which were modulated by low Pi treatment. Overall, we observed a wide variety of genetic variations in the REP, RRSR, RPPUE, and RPUpE among 157 rice accessions. The current GWAS analysis—which is thoroughly discussed below—detected 18 QTLs with 85 candidate genes.
Co-locations of the identified QTLs in previous studies
The co-locations of our observed QTLs with previously published QTLs related to Pi starvation were identified. Interestingly, our results showed some similarity with the previous studies, which confirms the reliability of our findings. Specifically, the qRPPUE12.18 for RPPUE located on chromosome 12 shared a similar region with a QTL, found by Wissuwa et al. (1998) for PUP and PUE which was flanked by the two markers G2140 and C443 and qRPPUE1.2 overlapped with a QTL, identified by Wang et al. (2014) for Pi translocation efficiency that was in the proximity of the BIN31-BIN32 markers on chromosome 1. qRPUpE1.5 shared a similar region with a QTL, also found by Wang et al. (2014) for the total above-ground Pi uptake which was flanked by the two BIN46-BIN47 markers on chromosome 1. qRRSR1.4 was close to QTLs, which were detected by Wang et al. (2014) flanked by marker RM283 (Xiang et al. 2015) and markers BIN33-BIN34 (Wang et al. 2014) for relative root dry weight, panicle number per plant, and Pi translocation efficiency.
It is worth noting that the common QTL qRPUUE9.16 was found for three traits, including REP, RPPUE, and RPUpE. Moreover, this QTL was found to be associated with the dry shoot weight, dry root weight, and total dry weight traits (Mai et al., 2020, under revision). These finding indicate that Pi uptake, Pi use efficiency, and REP were strongly correlated to the dry weight trait of rice plants.
Candidate genes for low Pi tolerance in the Vietnamese rice collection
Many mechanisms are activated in response to Pi starvation conditions in plants. Numerous studies have showed that protein kinases and phosphatases play crucial roles in signal transduction in response to stressors at the cellular level in plants (Sopory and Munshi 1998; Luan 1998; Wang et al. 2007). It is interesting to note that among the low Pi-responsive gene list, several genes encoding protein kinases (LOC_Os01g57420.1, LOC_Os01g57480.1, LOC_Os07g42940.1, LOC_Os09g12290.1, LOC_Os09g12300.1, and LOC_Os09g17010.1) and one gene encoding protein phosphatase (PP; LOC_Os10g39540.1) were found.
One very important gene, Os07g0622000 or OsSAPK2, which encodes for stress-activated protein kinase and belongs to the sucrose non-fermenting-1-related protein kinase 2 family (SnRK2s), was identified. This kinase plays a crucial role in energy sensing and in adaptive responses to stress in plants (Colina et al. 2019). OsSAPK2 was found to be located on chromosome 7 near the Dj07_25716591R marker and belongs to qRPPUE7.14. Recently, SnRK2s were reported to be negatively regulated by PP2C via dephosphorylation in the PP2C-SnRK2 complex (Hirayama and Umezawa 2010). Surprisingly, OsSAPK2 has also been shown to be involved in nutrient starvation. SnRK2.8, a homolog of OsSAPK2 in A. thaliana, was downregulated in the roots by Pi deprivation and consequently led to decrease in the growth of A. thaliana under Pi deprivation conditions (Umezawa et al. 2004; Shin et al. 2007). In rice, the expression level of OsSAPK2 was also markedly reduced from 6 to 24 h following the onset of Pi starvation conditions, which resulted from the reduction of the P content in rice grains (Lou et al. 2020). In addition, a number of studies have been carried out to demonstrate the function of this protein in plant defense mechanisms against biotic and abiotic stressors. For example, the expression level of OsSAPK2 was increased in response to Xanthomonas oryzae pv. Oryzae strain PXO99A (Hu et al. 2015) and Xanthomonas oryzae pv. oryzicola strain RS105. OsSAPK2 was also involved in the abscisic acid signal transduction pathway (Fujita et al. 2011), during drought, high salinity, and polyethylene glycol treatments in rice (Umezawa et al. 2004; Lou et al. 2017; Jadamba et al. 2020). Based on these findings, OsSAPK2 appears to be a promising gene for regulating stress response in rice.
Phosphatases (PPs) are hydrolytic enzymes that cleave a Pi group from its unavailable organic phosphates into assimilable Pi for plant uptake. Since 50–80% of the total Pi in agricultural and natural soils is presented in organic forms (Wang et al. 2009), PPs play an essential role in releasing Pi from these organic sources (Brinch-Pedersen et al. 2002). In plants, the serine/threonine PPs are divided into two types, PP1 and PP2, and PP2 is further divided into three groups, 2A, 2B, and 2C, based on their dependence on divalent cations (Ingebritsen and Cohen, 1983). A number of studies have demonstrated the function of these PPs in signal transduction, metabolism, and hormone regulation and stress response (Luan 1998; Schweighofer et al. 2004; Farkas et al. 2007). The current GWAS identified a PP2C gene, Os10g0541200 (LOC_Os10g39540), located on chromosome 10 near the Dj10_21135126R marker and co-located with qREP10.17. Various studies have been carried out to demonstrate the involvement of this gene in rice plants dealing with salt stress and abscisic acid treatment (Wang et al. 2016) as well as in regulating seed dormancy (Wu et al. 2016). Up till recently, there has not been any research showing the involvement of Os10g0541200 in the Pi starvation response; however, a recent study demonstrated the function of this gene in response to low ammonium nitrate treatment (Yang et al. 2017). Furthermore, a gene encoding protein PP2A in Zea mays L. was shown to be upregulated over 60 times in the roots following a 7 h Pi starvation (Wang et al. 2017). This result showed the role of PP2A in response to Pi starvation. Therefore, Os10g0541200 may be a potential candidate gene for improving plant tolerance to Pi deficiency.
Furthermore, the RNA-Seq transcriptome data was used to search for Pi-responsive genes among our list of candidate genes. In fact, the activation of Pi starvation-induced genes was regulated by a number of TFs, of which PHR1 is the key factor in vascular plants (Devaiah et al. 2007). In our study, although PHR1 was not found, the transcriptional data showed that two other TFs, including bHLH (LOC_Os03g59670.1) and NAC (LOC_Os12g29330.1), were found to be significantly upregulated in the roots and shoots of rice plants exposed to Pi starvation conditions (https://genevestigator.com). In addition, LOC_Os12g29330.1 was observed to be upregulated to response to osmotic stress (Dong et al. 2019) and Magnaporthe oryzae (Zhang et al. 2016). Among the list of candidate genes generated from the transcriptional data that were regulated by low Pi treatment, LOC_Os05g31670.1 named OsPM1 (O. sativa PLASMA MEMBRANE PROTEIN 1) was not only reported to be highly upregulated 214.41 times more in the roots of Nipponbare rice exposed to treated medium without iron compared to medium without both iron and P but also strongly responsive to drought in various tissues obtained from young seedlings to the leaves, roots, stems, and panicles (Yao et al. 2018) as well as to salt stress in the roots (Kong et al. 2019). The differential expression pattern from https://genevestigator.com also revealed another interesting Pi-responsive gene named OsCML15, a calcium signaling-related gene, which was downregulated 15.73 times more in the roots of a Pi-tolerant variety (Dular) compared to a Pi-sensitive one (Pusa Basmati 1) under low Pi supply. Notably, this gene also responded to salt stress by extensively increasing the expression over 100 times more under high salt treatment in rice (O. rufipogon Griff.) (Wang et al. 2017). OsCML15 was also stimulated by brown planthopper (Lv et al. 2014) and Xanthomonas strain PXO99A (Pérez-Quintero et al. 2013) as well as heat stress (Jung and An 2012). These interesting results corroborate our findings and suggest a new research question for validating the function of these genes in our Vietnamese rice collection.
In addition, glycerophosphoryl diester phosphodiesterase proteins (GDPDs) function in remobilizing glycerophosphate in cells, thus maintaining phosphate homeostasis (Corda et al. 2014). In our study, Os09g0339800, which is located in qRPUUE9.16 close to the Dj09_10404314F marker and named GDPD13, was found. Impressively, a number of studies have demonstrated highly induced expression levels of GDPDs under low Pi supply in Arabidopsis (Misson et al. 2005; Gaude et al. 2008), chickpea (Cicer arietinum L.) (Mehra and Giri 2016), maize (Z. mays L.) (Calderon-Vazquez et al. 2008), and white lupin (Lupinus albus L.) (Cheng et al. 2011) as well as in rice (Mehra and Giri 2016; Mehra et al. 2019). For instance, the expression levels of OsGDPD1 and OsGDPD5 were 42 and 67 times higher in low Pi conditions, respectively, compared to that upon full Pi supply after 7 days in low Pi medium. The expression levels increased further when rice plants were grown in low Pi medium for 15 days. Approximately 65 and 147 times higher expression levels of the OsGDPD1 and OsGDPD5 genes, respectively, were observed when plants were treated with a low Pi supply (Mehra and Giri 2016). GDPD13 was also found in the QTL for root weight, shoot weight, and total weight in our previous study (Mai et al., 2020, under revision). These expression profiles highlight the importance of GDPDs in plant adaptation to Pi starvation conditions and suggest further studies on GDPD13 to understand the function of this gene in acclimatization to Pi starvation.
Alternative mechanism to OsPSTOL1 in response to Pi starvation in rice
In this analysis, 62% of rice genotypes did not possess the OsPSTOl1 gene in their genome sequence; however, there was no significant difference between the two groups in three out of the four investigated traits under low Pi conditions until 6 weeks. Compared to the absence of OsPSTOl1, the presence of OsPSTOl1 did not result in higher growth. With respect to the REP trait, the accessions without OsPSTOl1 had higher REP than the ones with the gene. Thus, the hypothesis regarding OsPSTOL1 as the exclusive genetic factor for tolerance to Pi starvation in most global rice varieties does not match with our results. Therefore, other gene(s) derived from our GWAS analysis in the Vietnamese rice collection may show potential in rendering tolerance to the plant in response to Pi starvation. Further studies are required for the validation of these gene(s). These results also provide candidate gene(s) that may be used in cloning to improve rice adaptability to low Pi supply stress.
Conclusion
The ideal Pi starvation-tolerant genotype should have both high Pi uptake and use efficiency. Thus, in the present study, we used the Vietnamese rice collection to assess these processes and their associated values. Some promising QTLs (including qRPPUE7.14, qRRSR5.11, qREP10.17, and qRPUUE9.16) and candidate genes (OsSAPK2, OsGDPD13, OsPM19, OsCML15, and PP2C) were identified. Our findings provide some novel fundamental insights regarding various Pi deficiency response factors within the diverse genetic makeup of Vietnamese rice landraces. This valuable information may help to facilitate the genetic manipulation of rice to improve P deficiency resistance in the context of sustainable agriculture. Further studies should be carried out afterward to validate the promising candidate genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgment
We would like to thank Prof. Michel Lebrun for the financial support to multiply the rice collection, Dr. Mai Duc Chung, Dr. Phung Thi Phuong Nhung, Ms. Nguyen Van Anh, Ms. Ngo Le Na for their extensive help with experimental procedures and sample harvesting.
Author contributions
Nga T. P. MAI and Huong Thi Mai TO conceived and designed the experiments. Nga T. P. MAI, Van Hiep NGUYEN, Thi Quynh Anh CHU and Hanh Thi KIEU carried out the phenotyping experiments. Huong Thi Mai TO, Nga T. P. MAI and Khang Quoc LE performed bioinformatics analysis. Nga T. P. MAI and Huong Thi Mai TO wrote the paper. All authors read and approved the manuscript for publication.
Funding
This research was funded by the University of Science and Technology of Hanoi (USTH), Vietnam academy of Science and Technology (VAST) under Grant Number USTH.BIO.01/19–20 to Nga T.P. MAI.
Compliance with ethical standards
The author(s) declare that they have no conflict of interest.
All authors are sure that all data and materials as well as software application support our published claims and comply with field standards.
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huong Thi Mai To, Email: [email protected].
Khang Quoc Le, Email: moc.liamg@79gnahkcouqel.
Hiep Van Nguyen, Email: nv.ude.suh@75T_peiHnaVneyugN.
Linh Viet Duong, Email: moc.liamg@2149hnilteiv.
Hanh Thi Kieu, Email: [email protected].
Quynh Anh Thi Chu, Email: [email protected].
Trang Phuong Tran, Email: [email protected].
Nga T. P. Mai, Email: [email protected], Email: moc.liamg@agngnouhpiam.
References
- Ajmera I, Charlie Hodgman T, Lu C. An integrative systems perspective on plant phosphate research. Gen (Basel) 2019 10.3390/genes10020139. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Balemi T, Negisho K. Management of soil phosphorus and plant adaptation mechanisms to phosphorus stress for sustainable crop production: a review. J Soil Sci Plant Nutr. 2012;12:547–561. 10.4067/s0718-95162012005000015. [CrossRef] [Google Scholar]
- Brinch-Pedersen H, Sørensen LD, Holm PB. Engineering crop plants: getting a handle on phosphate. Trends Plant Sci. 2002;7:118–125. 10.1016/S1360-1385(01)02222-1. [Abstract] [CrossRef] [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant—and species-specific levels. J Exp Bot. 2008;59:2479–2497. 10.1093/jxb/ern115. [Abstract] [CrossRef] [Google Scholar]
- Chen W, Gao Y, Xie W, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46:714–721. 10.1038/ng.3007. [Abstract] [CrossRef] [Google Scholar]
- Cheng L, Bucciarelli B, Liu J, et al. White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol. 2011;156:1131–1148. 10.1104/pp.111.173724. [Abstract] [CrossRef] [Google Scholar]
- Colina F, Amaral J, Carbó M, et al. Genome-wide identification and characterization of CKIN/SnRK gene family in Chlamydomonas reinhardtii. Sci Rep. 2019;9:1–16. 10.1038/s41598-018-35625-8. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Corda D, Mosca MG, Ohshima N, et al. The emerging physiological roles of the glycerophosphodiesterase family. FEBS J. 2014;281:998–1016. 10.1111/febs.12699. [Abstract] [CrossRef] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;143:1789–1801. 10.1104/pp.106.093971. [Abstract] [CrossRef] [Google Scholar]
- Dong Z, Li W, Liu J, et al. The rice phosphate transporter protein OsPT8 regulates disease resistance and plant growth. Sci Rep. 2019;9:2–11. 10.1038/s41598-019-41718-9. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, et al. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007;12:169–176. 10.1016/j.tplants.2007.03.003. [Abstract] [CrossRef] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. 10.1007/s10265-011-0412-3. [Abstract] [CrossRef] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature. 2012;488:535–539. 10.1038/nature11346. [Abstract] [CrossRef] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, et al. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 2008;56:28–39. 10.1111/j.1365-313X.2008.03582.x. [Abstract] [CrossRef] [Google Scholar]
- Gilbert N. The disappearing nutrient. Nature. 2009;461:716–718. 10.1038/461716a. [Abstract] [CrossRef] [Google Scholar]
- Haefele SM, Nelson A, Hijmans RJ. Soil quality and constraints in global rice production. Geoderma. 2014;235–236:250–259. 10.1016/j.geoderma.2014.07.019. [CrossRef] [Google Scholar]
- Hammond JP, Broadley MR, White PJ, et al. Shoot yield drives phosphorus use efficiency in Brassica oleracea and correlates with root architecture traits. J Exp Bot. 2009;60:1953–1968. 10.1093/jxb/erp083. [Abstract] [CrossRef] [Google Scholar]
- Hirayama T, Umezawa T. The PP2c-SnRK2 complex: the central regulator of an abscisic acid signaling pathway. Plant Signal Behav. 2010;5:160–163. 10.4161/psb.5.2.10460. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hruz T, Laule O, Szabo G, et al. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform. 2008;2008:1–5. 10.1155/2008/420747. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hu DD, Zhang F, Huang LY, et al. Stress-activated protein kinase OsSAPK2 involved in regulating resistant response to Xanthomonas oryzae pv. oryzae in rice. Acta Agron Sin. 2015;41:1191–1200. 10.3724/SP.J.1006.2015.01191. [CrossRef] [Google Scholar]
- Ingebritsen TS, Cohen P. The Protein phosphatases involved in cellular regulation: 1. classification and substrate specificities. Eur J Biochem. 1983;132:255–261. 10.1111/j.1432-1033.1983.tb07357.x. [Abstract] [CrossRef] [Google Scholar]
- Jadamba C, Kang K, Paek NC, et al. Overexpression of rice expansin7 (Osexpa7) confers enhanced tolerance to salt stress in rice. Int J Mol Sci. 2020;21:454. 10.3390/ijms21020454. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jung KH, An G. Application of MapMan and RiceNet drives systematic analyses of the early heat stress transcriptome in rice seedlings. J Plant Biol. 2012;55:436–449. 10.1007/s12374-012-0270-0. [CrossRef] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, et al. Improvement of the oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:3–10. 10.1186/1939-8433-6-4. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kennelly M, O’Mara J, Rivard C, et al. Introduction to abiotic disorders in plants. Plant Heal Instr. 2012 10.1094/phi-i-2012-10-29-01. [CrossRef] [Google Scholar]
- Kirkby EA, Johnston AE. Soil and fertilizer phosphorus in relation to crop nutrition. In: White PJ, Hammond JP, editors. The ecophysiology of plant-phosphorus interactions. Springer, Dordrecht: Plant Ecophysiology; 2008. pp. 177–223. [Google Scholar]
- Kong W, Zhong H, Gong Z, et al. Meta-analysis of salt stress transcriptome responses in different rice genotypes at the seedling stage. Plants. 2019;8:64. 10.3390/plants8030064. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Le C, Zha Y, Li Y, et al. Eutrophication of lake waters in China: Cost, causes, and control. Environ Manage. 2010;45:662–668. 10.1007/s00267-010-9440-3. [Abstract] [CrossRef] [Google Scholar]
- Lekklar C, Suriya-arunroj D, Pongpanich M, et al. Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. Genes (Basel) 2019;10:1–20. 10.3390/genes10080562. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li J, Xie Y, Dai A, et al. Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using introgression lines of rice. J Genet Genom. 2009;36:173–183. 10.1016/S1673-8527(08)60104-6. [Abstract] [CrossRef] [Google Scholar]
- Li Y, Zhang J, Zhang X, et al. Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 2015;230:23–32. 10.1016/j.plantsci.2014.10.001. [Abstract] [CrossRef] [Google Scholar]
- Lindsay WL. Chemical equilibria in soils. New York: Wiley-Interscience; 1979. [Google Scholar]
- Liu X, Zhao X, Zhang L, et al. TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct Plant Biol. 2013;40:329–341. 10.1071/FP12242. [Abstract] [CrossRef] [Google Scholar]
- Lizbeth D, ´ Opez-Arredondo L, Leyva-González MA,, et al. Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol. 2014;65:95–123. 10.1146/annurev-arplant-050213-035949. [Abstract] [CrossRef] [Google Scholar]
- Lou D, Chen Z, Yu D, Yang X. SAPK2 contributes to rice yield by modulating nitrogen metabolic processes under reproductive stage drought stress. Rice. 2020;13:35. 10.1186/s12284-020-00395-3. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lou D, Wang H, Liang G, Yu D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front Plant Sci. 2017;8:993. 10.3389/fpls.2017.00993. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Luan S. Protein phosphatases and signaling cascades in higher plants. Trends Plant Sci. 1998;3:271–275. 10.1016/S1360-1385(98)01258-8. [CrossRef] [Google Scholar]
- Lv W, Du B, Shangguan X, et al. BAC and RNA sequencing reveal the brown planthopper resistance gene BPH15 in a recombination cold spot that mediates a unique defense mechanism. BMC Genom. 2014;15:1–16. 10.1186/1471-2164-15-674. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- MacDonald GK, Bennett EM, Potter PA, Ramankutty N. Agronomic phosphorus imbalances across the world’s croplands. Proc Natl Acad Sci U S A. 2011;108:3086–3091. 10.1073/pnas.1010808108. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. 10.1056/NEJMra0905980. [Abstract] [CrossRef] [Google Scholar]
- Mehra P, Giri J. Rice and chickpea GDPDs are preferentially influenced by low phosphate and CaGDPD1 encodes an active glycerophosphodiester phosphodiesterase enzyme. Plant Cell Rep. 2016;35:1699–1717. 10.1007/s00299-016-1984-0. [Abstract] [CrossRef] [Google Scholar]
- Mehra P, Pandey BK, Verma L, Giri J. A novel glycerophosphodiester phosphodiesterase improves phosphate deficiency tolerance in rice. Plant Cell Environ. 2019;42:1167–1179. 10.1111/pce.13459. [Abstract] [CrossRef] [Google Scholar]
- Miao J, Sun J, Liu D, et al. Characterization of the promoter of phosphate transporter TaPHT1.2 differentially expressed in wheat varieties. J Genet Genom. 2009;36:455–466. 10.1016/S1673-8527(08)60135-6. [Abstract] [CrossRef] [Google Scholar]
- Misson J, Raghothama KG, Jain A, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci U S A. 2005;102:11934–11939. 10.1073/pnas.0505266102. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002;31:341–353. 10.1046/j.1365-313X.2002.01356.x. [Abstract] [CrossRef] [Google Scholar]
- Mukherjee A, Sarkar S, Chakraborty AS, et al. Phosphate acquisition efficiency and phosphate starvation tolerance locus (PSTOL1) in rice. J Genet. 2014;93:683–688. 10.1007/s12041-014-0424-6. [Abstract] [CrossRef] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 1980;8:4321–4326. 10.1093/nar/8.19.4321. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nagarajan VK, Jain A, Poling MD, et al. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011;156:1149–1163. 10.1104/pp.111.174805. [Abstract] [CrossRef] [Google Scholar]
- Neto AP, Favarin JL, Hammond JP, et al. Analysis of phosphorus use efficiency traits in coffea genotypes reveals coffea arabica and coffea canephora have contrasting phosphorus uptake and utilization efficiencies. Front Plant Sci. 2016;7:408. 10.3389/fpls.2016.00408. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nilsson L, MÜller R, Nielsen TH, Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant, Cell Environ. 2007;30:1499–1512. 10.1111/j.1365-3040.2007.01734.x. [Abstract] [CrossRef] [Google Scholar]
- Ozturk L, Eker S, Torun B, Cakmak I. Variation in phosphorus efficiency among 73 bread and durum wheat genotypes grown in a phosphorus-deficient calcareous soil. Plant Soil. 2005;269:69–80. 10.1007/s11104-004-0469-z. [CrossRef] [Google Scholar]
- Pariasca-Tanaka J, Chin JH, Dramé KN, et al. A novel allele of the P-starvation tolerance gene OsPSTOL1 from African rice (Oryza glaberrima Steud) and its distribution in the genus Oryza. Theor Appl Genet. 2014;127:1387–1398. 10.1007/s00122-014-2306-y. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Pérez-Quintero AL, Rodriguez-R LM, Dereeper A, et al. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae Strains. PLoS ONE. 2013;8:e68464. 10.1371/journal.pone.0068464. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Phung NTP, Mai CD, Mournet P, et al. Characterization of a panel of Vietnamese rice varieties using DArT and SNP markers for association mapping purposes. BMC Plant Biol. 2014;14:1–16. 10.1186/s12870-014-0371-7. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. 10.1146/annurev.arplant.50.1.665. [Abstract] [CrossRef] [Google Scholar]
- Rice EW, Baird RB, Eaton AD (2017) Standard methods for the examination of water and wastewater, 23rd Edition. In: Am. Public Heal. Assoc. Am. Water Work. Assoc. Water Environ. Fed
- Schünmann PHD, Richardson AE, Smith FW, Delhaize E. Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.) J Exp Bot. 2004;55:855–865. 10.1093/jxb/erh103. [Abstract] [CrossRef] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. 10.1016/j.tplants.2004.03.007. [Abstract] [CrossRef] [Google Scholar]
- Seo HM, Jung Y, Song S, et al. Increased expression of OsPT1, a high-affinity phosphate transporter, enhances phosphate acquisition in rice. Biotechnol Lett. 2008;30:1833–1838. 10.1007/s10529-008-9757-7. [Abstract] [CrossRef] [Google Scholar]
- Shimizu A, Kato K, Komatsu A, et al. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.): Fine QTL mapping and multivariate analysis of related traits. Theor Appl Genet. 2008;117:987–996. 10.1007/s00122-008-0838-8. [Abstract] [CrossRef] [Google Scholar]
- Shin R, Alvarez S, Burch AY, et al. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc Natl Acad Sci U S A. 2007;104:6460–6465. 10.1073/pnas.0610208104. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sopory SK, Munshi M. Protein kinases and phosphatases and their role in cellular signaling in plants. CRC Crit Rev Plant Sci. 1998;17:245–318. 10.1080/07352689891304230. [CrossRef] [Google Scholar]
- Stigter KA, Plaxton WC. Molecular mechanisms of phosphorus metabolism and transport during leaf senescence. Plants. 2015;4:773–798. 10.3390/plants4040773. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sun S, Gu M, Cao Y, et al. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012;159:1571–1581. 10.1104/pp.112.196345. [Abstract] [CrossRef] [Google Scholar]
- Taiz L, Zeiger E (2002) Plant physiology. 3rd edn. Ann Bot. 91:750–751. 10.1093/aob/mcg079
- To HTM, Nguyen HT, Dang NTM, et al. Unraveling the genetic elements involved in shoot and root growth regulation by Jasmonate in rice using a genome-wide association study. Rice. 2019 10.1186/s12284-019-0327-5. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, et al. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:17306–17311. 10.1073/pnas.0407758101. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157:423–447. 10.1046/j.1469-8137.2003.00695.x. [Abstract] [CrossRef] [Google Scholar]
- Vigueira CC, Small LL, Olsen KM. Long-term balancing selection at the phosphorus starvation tolerance 1 (PSTOL1) locus in wild, domesticated and weedy rice (Oryza) BMC Plant Biol. 2016;16:1–10. 10.1186/s12870-016-0783-7. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang H, Chevalier D, Larue C, et al. The protein phosphatases and protein kinases of arabidopsis thaliana. Arab B. 2007 10.1199/tab.0106. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang J, Sun J, Miao J, et al. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot. 2013;111:1139–1153. 10.1093/aob/mct080. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang K, Cui K, Liu G, et al. Identification of quantitative trait loci for phosphorus use efficiency traits in rice using a high density SNP map. BMC Genet. 2014;15:1–15. 10.1186/s12863-014-0155-y. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang S, Cao M, Ma X, et al. Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front Plant Sci. 2017;8:1427. 10.3389/fpls.2017.01427. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang WS, Zhao XQ, Li M, et al. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J Exp Bot. 2016;67:405–419. 10.1093/jxb/erv476. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang X, Wang Y, Tian J, et al. Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol. 2009;151:233–240. 10.1104/pp.109.138891. [Abstract] [CrossRef] [Google Scholar]
- Wissuwa M, Ae N. Further characterization of two QTLs that increase phosphorus uptake of rice Oryza sativa L under phosphorus deficiency. Plant Soil. 2001;237(2):275–286. 10.1023/A:1013385620875. [CrossRef] [Google Scholar]
- Wissuwa M, Kondo K, Fukuda T, et al. Unmasking novel loci for internal phosphorus utilization efficiency in rice germplasm through genome-wide association analysis. PLoS ONE. 2015 10.1371/journal.pone.0124215. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wissuwa M, Yano M, Ae N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.) Theor Appl Genet. 1998;97:777–783. 10.1007/s001220050955. [CrossRef] [Google Scholar]
- Wu T, Yang C, Ding B, et al. Microarray-based gene expression analysis of strong seed dormancy in rice cv. N22 and less dormant mutant derivatives. Plant Physiol Biochem. 2016;99:27–38. 10.1016/j.plaphy.2015.12.001. [Abstract] [CrossRef] [Google Scholar]
- Xiang C, Ren J, Zhao XQ, et al. Genetic dissection of low phosphorus tolerance related traits using selected introgression lines in rice. Rice Sci. 2015;22:264–274. 10.1016/j.rsci.2015.05.020. [CrossRef] [Google Scholar]
- Yang HC, Kan CC, Hung TH, et al. Identification of early ammonium nitrate-responsive genes in rice roots. Sci Rep. 2017;7:16885–16885. 10.1038/s41598-017-17173-9. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yang W, Guo Z, Huang C, et al. Genome-wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J Exp Bot. 2015;66:5605–5615. 10.1093/jxb/erv100. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yao L, Cheng X, Gu Z, et al. The AWPM-19 family protein OsPM1 mediates abscisic acid influx and drought response in rice. Plant Cell. 2018;30:1258–1276. 10.1105/tpc.17.00770. [Abstract] [CrossRef] [Google Scholar]
- Yoshida S, Forno DA, Cock J (1971) Laboratory Manual for Physiological Studies of Rice.61
- Yumnam JS, Rai M, Tyagi W. Allele mining across two low-P tolerant genes PSTOL1 and PupK20-2 reveals novel haplotypes in rice genotypes adapted to acidic soils. Plant Genet Resour Charact Util. 2017;15:221–229. 10.1017/S1479262115000544. [CrossRef] [Google Scholar]
- Zhang F, Wu X-N, Zhou H-M, et al. Overexpression of rice phosphate transporter gene OsPT6 enhances phosphate uptake and accumulation in transgenic rice plants. Plant Soil. 2014;384:259–270. 10.1007/s11104-014-2168-8. [CrossRef] [Google Scholar]
- Zhang Y, Zhao J, Li Y, et al. Transcriptome analysis highlights defense and signaling pathways mediated by rice pi21 gene with partial resistance to Magnaporthe oryzae. Front Plant Sci. 2016;7:1834. 10.3389/fpls.2016.01834. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from Physiology and Molecular Biology of Plants are provided here courtesy of Springer
Full text links
Read article at publisher's site: https://doi.org/10.1007/s12298-020-00902-2
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc7688854?pdf=render
Citations & impact
Impact metrics
Article citations
Rice breeding for low input agriculture.
Front Plant Sci, 15:1408356, 21 Jun 2024
Cited by: 0 articles | PMID: 38974981
Review
Genome-wide association study reveals useful QTL and genes controlling the fatty acid composition in rice bran oil using Vietnamese rice landraces.
Funct Integr Genomics, 23(2):150, 09 May 2023
Cited by: 0 articles | PMID: 37156920
Deep polygenic neural network for predicting and identifying yield-associated genes in Indonesian rice accessions.
Sci Rep, 12(1):13823, 15 Aug 2022
Cited by: 1 article | PMID: 35970979 | PMCID: PMC9378700
Genomic regions and candidate genes selected during the breeding of rice in Vietnam.
Evol Appl, 15(7):1141-1161, 09 Jul 2022
Cited by: 3 articles | PMID: 35899250 | PMCID: PMC9309459
Prospects of genetics and breeding for low-phosphate tolerance: an integrated approach from soil to cell.
Theor Appl Genet, 135(11):4125-4150, 07 May 2022
Cited by: 6 articles | PMID: 35524816 | PMCID: PMC9729153
Review Free full text in Europe PMC
Go to all (7) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Discovery of new genetic determinants of morphological plasticity in rice roots and shoots under phosphate starvation using GWAS.
J Plant Physiol, 257:153340, 15 Dec 2020
Cited by: 7 articles | PMID: 33388665
Characterization of contrasting rice (Oryza sativa L.) genotypes reveals the Pi-efficient schema for phosphate starvation tolerance.
BMC Plant Biol, 21(1):282, 21 Jun 2021
Cited by: 16 articles | PMID: 34154533 | PMCID: PMC8215752
Genome-wide Association Study of a Panel of Vietnamese Rice Landraces Reveals New QTLs for Tolerance to Water Deficit During the Vegetative Phase.
Rice (N Y), 12(1):4, 28 Jan 2019
Cited by: 23 articles | PMID: 30701393 | PMCID: PMC6357217
Phenotypes and Molecular Mechanisms Underlying the Root Response to Phosphate Deprivation in Plants.
Int J Mol Sci, 24(6):5107, 07 Mar 2023
Cited by: 1 article | PMID: 36982176 | PMCID: PMC10049108
Review Free full text in Europe PMC
Funding
Funders who supported this work.
University of Science and Technology of Hanoi (USTH), Vietnam Academy of Science and Technology (1)
Grant ID: USTH.BIO.01/19-20






