Abstract
Free full text

microRNAs Promoting Growth of Gastric Cancer Xenografts and Correlation to Clinical Prognosis
Abstract
The annual death toll for gastric cancer is in the range of 700,000 worldwide. Even in patients with early-stage gastric cancer recurrence within five years has been observed after surgical resection and following chemotherapy with therapy-resistant features. Therefore, the identification of new targets and treatment modalities for gastric cancer is of paramount importance. In this review we focus on the role of microRNAs with documented efficacy in preclinical xenograft models with respect to growth of human gastric cancer cells. We have identified 31 miRs (-10b, -19a, -19b, -20a, -23a/b, -25, -27a-3p, -92a, -93, -100, -106a, -130a, -135a, -135b-5p, -151-5p, -187, -199-3p, -215, -221-3p, -224, -340a, -382, -421, -425, -487a, -493, -532-3p, -575, -589, -664a-3p) covering 26 different targets which promote growth of gastric cancer cells in vitro and in vivo as xenografts. Five miRs (miRs -10b, 151-5p, -187, 532-3p and -589) additionally have an impact on metastasis. Thirteen of the identified miRs (-19b, -20a/b, -25, -92a, -106a, -135a, -187, -221-3p, -340a, -421, -493, -575 and -589) have clinical impact on worse prognosis in patients.
Gastric cancer (GC) is the third-leading cause of cancer worldwide and is the forth most common cancer with an annual death toll of 700,000 worldwide (1). Intestinal-type and diffuse-type are the major histological subtypes (2). A total of 90% of GCs are adenocarcinomas which arise from the glandular epithelium (1). A total of 1-3% of GCs are hereditary cancers due to inactivating mutations in E-cadherin (1). From a molecular point of view, the following subtypes have been defined: Epstein-Barr-Virus (EBV)-positive with pronounced DNA hypermethylation, genomic stable subtype with distinctive genomic alterations, the microsatellite instability subtype (MSI) and the chromosomal instability (CIN) subtype characterized by aneuploidy and focal amplification of transmembrane receptor tyrosine kinases (3). These subtypes are correlated with different survival and outcomes of chemotherapy (3). Several agents have been approved for the treatment of gastric cancer: chemotherapy-based drugs such as 5-fluorouracil (5-FU)-related agents, docetaxel, doxorubicin and mitomycin B as well as monoclonal antibodies (mABs) such as trastuzumab (HER2 as a target) for HER2-positive patients, ramicurumab (VEGFR2 as a target) and checkpoint-inhibitory antibody pembrolizumab directed against programmed cell death 1 (PD1) (5-10). Nevertheless, in patients with early-stage GC, recurrence within 5 years after surgical resection and subsequent chemotherapy has been observed (4,5). The prognosis of patients with advanced GC is significantly worse (4,5).
Therefore, the identification of new targets and treatment modalities is an important issue. In this review, we describe the role of microRNAs (miRs) in pathogenesis and metastasis of GC. We focus on miRs which are up-regulated in GC tissue in comparison to matching normal tissues which exhibit efficacy in preclinical in vivo models. We have excluded: miRs playing a functional role in Helicobacter pylori-related gastric cancer models, miRs exerting their function by interaction with non-coding RNAs, miRs delivered by exosomes and those induced in tumor cells by stromal cells.
microRNAs and Their Role in Oncology
miRs are double-stranded non-coding RNAs comprising 22-25 nucleotides (nts) which are generated from precursors transcribed in the nucleus, processed and finally 22-25 nts miR complexes are released into the cytoplasm (12-14). The guide strand is maintained and the other strand, the passenger strand, is degraded (13,14). A single miR species may repress several miRs and each mRNA can be degraded by several different miRs indicating their potential to modulate serveral pathways and cellular networks (15). miRs can be transcribed from single transcriptional units or from multicistronic transcripts from a coding gene, introns or non-coding genes (15).
In oncology, miRs function as oncogenes as well as tumor suppressors (TS), depending on the cell-type in which they are expressed (15-17). Genetic deletion of the miR-15/16 genetic locus in mice recapitulated the features of human chronic lymphocytic leukemia, supporting their role as tumor suppressors (18). An oncogenic role was identified for miR-221 which induced hepatocellular carcinomas in transgenic mice after liver-specific expression (19). miRs are also involved in metastasis. We have recently summarized their role in metastasis in breast-, prostate, ovarian, lung and pancreatic cancer (20).
miRs Inhibiting Tumor Suppressors
PTEN. miRs-221-3p (25), -382 (26), -425 (27) and -575 (28) (Figure 1) target phosphatase and tensin homolog PTEN which dephosphorylates phosphatidylinositol-3,4,5 triphosphate and thus negatively regulates the AKT signaling pathway (29,30). miRs -382 and -425 are induced by hypoxia-inducible factor 1 (HIF-1) or interleukin 1β (Il1β), respectively (26,27). miRs-221-3p, -382, -425 and -575 affect tumor growth of gastric cancer cell lines SNU-1, MKN1, NCI-N87 and MGC-803 respectively after subcutaneous implantation into nude mice (25-28). miR-382 also affects angiogenesis, by promoting sprouting, branching and network formation of blood vessels induced by vascular endothelial growth factor (VEGF) (26). Overexpression of miRs-221-3p and -575 in GC patients in comparison to matching normal tissues correlates with worse prognosis (25,28). All the discussed miRs activate AKT signaling. Data derived from The Cancer Genome Atlas (TGCA) show that miR-221 is overexpressed in GC tissues in comparison to matching normal tisses (Figure 2).
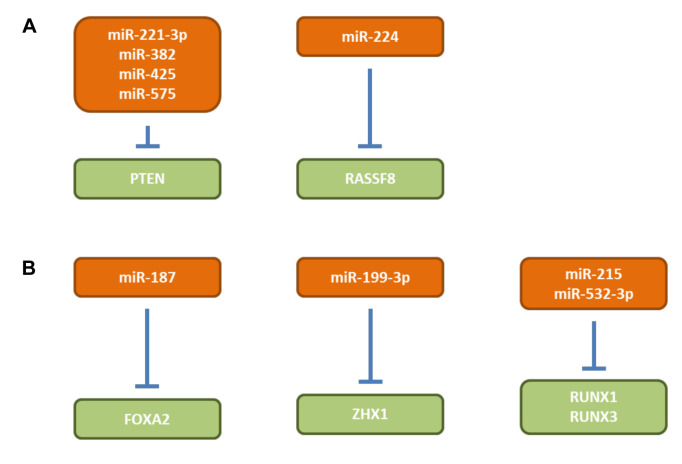
RASSF8. miR-224 (Figure 1) is induced by hypoxia and increases cell viability and invasion in gastric cancer cell lines SGC-7901 and MGC-803 (31). SCG-7901 cells transfected with a miR-224 antagomir exhibit reduced growth after subcutaneous implantation into nude mice (31). Ras-association domain containing protein 8 (RASSF8) was identified as a direct target of miR-224 (32). RASSF8 is a member of a family of 10 genes which are linked to processes such as cell proliferation, cell death and responses to hypoxia (32). The above mentioned in vitro effects were recapitulated by overexpression of RASSF8 in SGC-7901 and MGC-803 cells (31). RASSF8 overexpression inhibits nuclear factor ĸB (NFĸB) signaling and it functions as a TS (33,34).
microRNAs Targeting Transcription Factors
miR-187 (FOXA2). Increased expression of miR-187 (Figure 1) is associated with clinico-pathological features and prognosis in GC patients (35). miR-187 promotes proliferation, migration and invasion of SGC-7901 GC cells (35). SGC-7901 cells overxpressing miR-18 exhibit increased tumor growth (TG) after subcutaneous implantation into nuce mice and increased lung colonization after tail vein injection (35). Forkhead box protein A2 (FOXA2) was identified as a direct target of miR-187 (35). It was shown that FOXA2 mediates the biological functions of miR-187 (35). FOXA2 is a transcriptional factor which suppresses gastric carcinogenesis in vitro and in vivo (36).
miR-199-3p (ZHX1). miR-199-3p increases proliferation and suppresses apoptosis in vitro and in vivo of GC cell lines SGC-7901 and NCI-N87 (37) (Figure 1). Transcription factor zinc fingers and homeoboxes (ZHX1) was identified as a target for miR-199-3p (37). Restoring ZHX1 expression in SGC-7901/miR-199-3p cells inhibits cell proliferation induced by miR-199a-3p (37). It has been shown that ZHX1 inhibits GC cell growth through inducing cell-cycle arrest and apoptosis (38). The ZHX family consists of three members containing two Cys2His2 zinc finger domains and five homeobox DNA-binding motifs (39).
RUNX1 and RUNX3. miRs-215 and -532-3p (Figure 1) target runt-related transcription factors RUNX1 or RUNX3 respectively (40,41). Ectopic expression of miR-215 promotes migration and invasion of GES-1 and HGC-27 cells (35). RUNX1 could partially reverse the function of miR-215 (35). HGC-27 cells stably expressing miR-215 metastasize to the liver after intraperitoneal injection (40). miR-532-5p promotes growth and migration of BGC-823 gastric cancer cells (36). In vivo miR-532-3p promotes lung colonization after tail vein injection of BCG-823 gastric cancer cells transfected with miR-532-3p (41). The RUNX familiy of transcription factors are frequently inactivated in cancer and function as TS (42). RUNX1 plays a role in early steps of haematopoesis and acts as a TS in breast cancer (43). RUNX3 was shown to have a tumor- and metastasis suppressive role in GC (44-47).
miRs Targeting Ubiquitin-related Enzymes
miR-25. miR-25 (Figure 3A) is overexpressed in primary tumor tissues of GC patients and is significantly correlated with a more aggressive phenotype of GC in patients (48). miR-25 mediates proliferation, invasion and migration of GC in vitro and promotes TG in vivo (48). F-box/WD repeat containing protein 7 (FBXW7) was identified as a direct target of miR-25 (48). Restauration of expression of FBXW7 led to reversion of the described in vitro effects (48). F-box proteins constitute one of the four subunits of the ubiquitin-protein ligase system (49,50). FBXW7 has been shown to function as a TS of human tumorigenesis (51). Data from TCGA show that miR-25 is over-expressed in GC tissues in comparison to matching normal tissues (Figure 2).
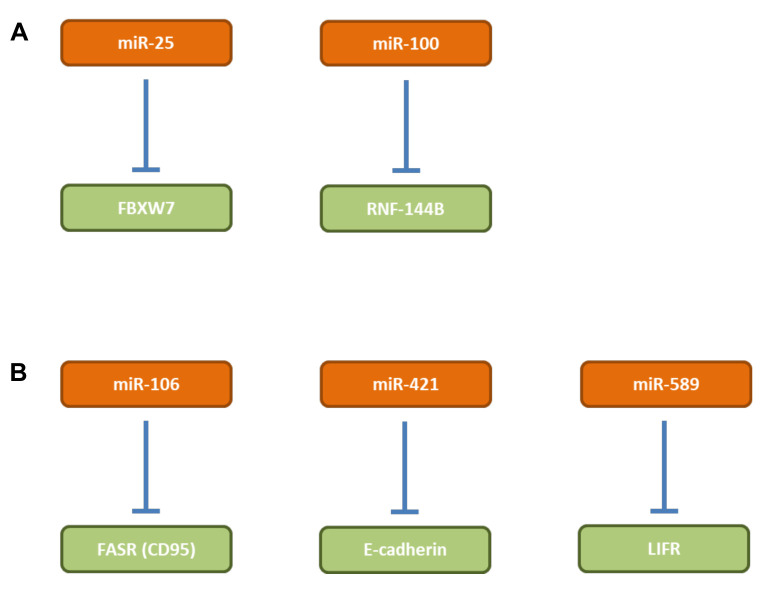
miR-100. Expression of miR-100 (Figure 3A) correlates with GC tumorigenesis and progression (52). Inhibition of miR-100 results in apoptosis of poorly differentiated gastric cancer cells, but not of non-cancerous gastric cells in vitro and in vivo (52). RNF144B, an E3-ubiquitin ligase was identified as a direct target of miR-100. RNF144B contains a single transmembrane domain close to the C-terminus, is localized in the nucleoli and mitochondria and promotes apoptosis under various cell damaging events by degradation of p53 (53-55).
miRs Targeting Transmembrane Receptors
miR-106a. miR-106a (Figure 3B) expression is significantly associated with tumor size, lymphatic and distant metastasis (56). miR-106 inhibits apoptosis of GC cells AGS, N87 and BGC823 (56,57). FASR (CD95) and caspase 3 were identified as direct targets of miR-106 (56,57). MicroRNA-106a functions as an oncogene in human gastric cancer and contributes to proliferation and metastasis in vitro and in vivo. miR-106 inhibits GC cell apoptosis through a FASR (CD95)-mediated, extrinsic cell death pathway (56-59). Data derived from TCGA show that miR-106a is over-expressed in GC tissues in comparison to matching normal tissues.
miR-421. miR-421 (Figure 3B) is increased in GC and is associated with poor prognosis (60). An antagomir of 421 reduces TG of subcutaneously implanted SGC-790/1 cells in nude mice (60). E-Cadherin and caspase 3, key regulators in the process of epithelial mesenchymal transition (EMT) have been identified as targets of miR-421 (60-62). Data derived from TCGA show that miR-421 is over-expressed in GC tissues in comparison to matching normal tissues.
miR-589. miR-589 (Figure 3B) is up-regulated in GC tissues and associated with poor prognosis in patients with GC (63). miR-589 induces migration and invasion of GC cell lines MGC803 and BCG823 in vitro (63). Endogenous overexpression of miR-589 promotes lung metastasis of MGC-803 cells after tail vein injection (63). Leukemia inhibitory factor receptor (LIFR) was identified as a direct target of miR-589 (63). LIFR suppresses the in vitro effects of miR-589 as described above (63). It was shown that LIFR is essential for miR-589-mediated promotion of PI3K/AKT signaling activation (63). LIFR (CD118) is a low-affinity subunit of the LIFR and together with a high-affinity converter subunit binds leukemia inhibitory factor (LIF) with high affinity. LIF is an interleukin 6 (IL6) class cytokine (64). LIFR has been identified as a metastasis suppressor in hepatocellular carcinoma (65). Data derived from TCGA atlas confirm that miR-589 is over-expressed in GC tissues in comparison to corresponding normal tissues (Figure 2).
miRs Involved in Signaling Pathways
miR-19a. mi-19a (Figure 4) promotes proliferation and tumorigenicity of MGC-803 and SGC-7901 GC cells (66). In vivo, increased TG was noted with SGC-7901 cells overexpressing miR-19a after subcutaneous implantation (66). Suppressor of cytokine signaling-1 (SOCS-1) was identified as a direct target of miR-19a (66). miR-19a expression levels are inversely correlated with SOCS-1 levels in GC tissues (66). SOCS-1 is member of a family of eight proteins each of which contains a src hmology 2 (SH2) domain and a COOH-terminal SOCS-box (67). SOCS-1 functions as an inhibitor of the janus tyrosine kinase (JAK) and signal transducer and activator of transcription 3 (STAT3) pathways (68,69).
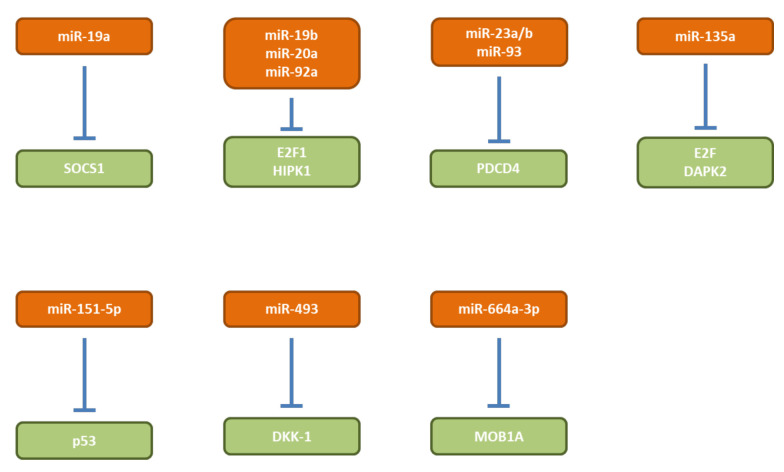
miR-19b, -20a, 92a. miRs-19b, -20a and 92a (Figure 4) are members of the mir-17-92 cluster and are overexpressed in human GC stem cells and are negatively correlated with the survival of GC patients (70). SGC7901 cells transduced with miRs-19b, -20a or -92a show improved TG after subcutaneous implantation into nude mice (70). Transcription factor E2F1 and homeodomain-interacting protein kinase (HIPK) were identified as targets of the miRs of the miR-17-92 cluster which are involved in self-renewing of GC stem cells (70). E2F can activate transcription of β-catenin-interacting protein (ICAT), an inhibitor of β-catenin (71). HIPK1 phosphorylates homeodomain transcription factors and co-repressors of homeobox transcriptions factors and modulates β-catenin signaling by interaction with dishevelled, a multi-domain scaffold protein required for virtually all WNT signaling activities (72). Data derived from the TCGA show that miRs-19b, -20a and -92a are overexpressed in GC tissues in comparison to corresponding normal tissues (Figure 2).
miR-493. miR-493 (Figure 4) is up-regulated in GC and promotes proliferation of SGC-709 and MGC-803 GC cells (73). TG of MGC-803 GC cells overexpressing miR-493 is increased after subcutaneous implantation in comparison to the control cell line (73). Dickkopf-related protein-1 (DKK-1) has been identified as direct target of miR-493 (73). Ectopic expression of DKK-1 can rescue the effect of a miR-493 inhibitor on proliferation in vitro and TG in vivo (73). DKK-1 is an antagonist of the WNT/β-catenin signaling pathway by isolating the low-density lipoprotein receptor-related protein 6 (LRP6) co-receptor so that it can not aid in activating the WNT signaling pathway (74,75). High DKK-1 expression regardless of β-catenin positivity is a crucial prognostic factor for predicting tumor recurrence and survival of patients with resected advanced GC (76). Data derived from TCGA show that miR-493 is over-expressed in GC tissues in comparison to corresponding normal tisses (Figure 2).
miR-664a-3p. miR-664a-3p (Figure 4) is up-regulated in GC tissues and promotes GC cell proliferation (77). miR-664a-3p mimics promote tumor volume of SGC790 and HGC27 GC tumor cells subcutaneously injected into nude mice, wheras a miR-664a-3p inhibitor decreases the tumor volume (77). MOB kinase activator 1A (MOB1A) was identified as a direct target of miR-664a-3p (77). MOB1A is a component of the Hippo pathway which is involved in restraining cell proliferation and promoting apoptosis. The Hippo pathway is frequently deregulated in different human cancers, but most Hippo pathway genes are not commonly mutated (78-80). MOB1A inhibits large tumor suppressor kinases 1 and 2 (LATS 1/2) which modulates two other kinases, nuclear dbf2-related 1 and 2 (NDR1 and NDR2) resulting in enrichment of yes-associated protein-1 (YAP) and transcriptional activator with PDZ-binding motif (TAZ) in the nucleus leading to a negative impact on transcription of Hippo pathway-related genes (78-80).
micro-RNAs Modulating Apoptosis
miRs-23a/b, -93. miRs-23a/b and -93 (Figure 4) are up-regulated in GC tissues in comparison to normal tissues (81,82). Programmed cell death 4 (PDCD4) has been identified as a direct target of miRs-23a/b and -93 (81,82). Introduction of miR-23a/b mimetics into MKN-45 and AGD GC cells reduces PDCD4 levels, whereas miR-23a/b antisense oligonucleotides increase PDCD4 levels (81). AGS cells transfected with PDCD4 small interfering RNA (siRNA) showed decreased apoptosis, transfection of a PDCD4 expression plasmid had the opposite effect (82). In vivo, MKN-45 and AGS GC cells overexpressing miR-23a/b or miR-93 respectively, exhibited increased tumor size and weight after subcutaneous implantation into immuno-compromized mide (81,82). PDCD4 mediates sensitivity to apoptosis by suppressing FLICE inhibitory protein (FLIP), a negative regulator of apoptosis (83,84). Down-regulation of PDCD4 has been observed in tumorigenesis and progression of human digestive cancers (85). In gastric GC, PDCD4 has been identified as a TS gene (86).
miR-135a. mR-135a (Figure 4) is up-regulated in GC patients and is associated with poor prognosis (87). miR-135a correlates with resistance to oxaliplatin (OXA) (87). OXA-resistant GC cells proliferate in response to miR-135a expression (87). TG of MGC-803/OXA GC transfected with miR-135a was increased and TG was reduced with MGC-803/OXA cells transfected with a miR-13 functional inhibitor after subcutaneous implantation into nude mice (87). E2F1 and death-associated protein kinase 2 (DAPK2) were identified as direct targets of miR-135a, promoting OXA-resistance (87). DAPK2 is stimulated by E2F1 and induces apoptosis by the kruppel-like factor (KLF/Sp1) transcription factor pathway (88). Restoration of DAPK2 TS in cancer cells by fusion protein containing a tumor-targeting module and DAPK2 reconstitution is a therapeutic strategy to selectively induce apoptosis in cancer cells (89,90). In patients with GC hypermethylation of the DAPK gene has been observed (91).
miR-151-5p. miR-151-5p (Figure 4) expression is increased by activated Notch1 pathway (92). miR-151 promotes growth of SC-M1 GC cells and induces focal adhesion kinase (FAK) (92). SC-M1 cells overexpressing miR-151-5p exhibit augmented tumor sizes in comparision to control cells after subcutaneous implantation into nude mice (92). After tail vein injection of these cells, metastatic nodules in the lungs were significantly increased (92). p53 was identified as a target of miR-151-5p (92). The apoptosis-mediating function of p53 in cancer is well documented (93,94). The suppressive effects of miR-151 antagomirs on Notch1 signalling-induced metastasis of SC-M1 cells could be partially recovered by cotransfection of an siRNA vector against p53 (92).
microRNAs Interfering With the Cell-Cycle
miR-27a-3p. miR-27a-3p (Figure 5A) is over-expressed in GC tissues and promotes proliferation and TC growth in NCI-N87, MGC-803 and GES-1 GC cells (95). Ectopic expression of miR-27a-3p in GES-1 cells promotes tumor weight and volume after subcutaneous administration into immuno-deficient mice, the opposite holds true for MGC-803 cells with inhibited expression of miR-27a-3p (95). B-cell translocation gene (BTG2) has been identified as a direct target of miR-27a-3p (95). BTG2 induces apoptosis and triggers cell cycle arrest by inhibiting G1/S transition (96,97). BTG2 is frequently deleted or mutated in B-cell malignancies and functions as a TS in several types of tumors (98).
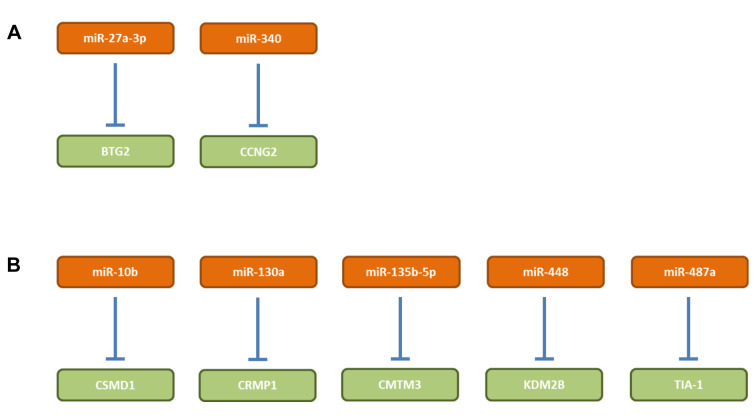
miR-340. miR-340 (Figure 5A) is up-regulated in GC tissues and cell lines (99). Elevated expression of miR-340 is correlated with adverse clinicopathological features and poor prognosis in GC patients (99). miR-340 promotes motility, proliferation and cell-cycle progression of GC cells such as SGC-7901 and MGC-803 (99). Inhibition of miR-340 decreased TG of MGC-803 cells in nude mice and cyclin G2 was identified as a direct target of miR-340 (99). CCNG2 is critical for the functional effects of miR-340 in GC cells, because knockdown of CCNG2 abrogated the effects of miR-340 knockdown in MGC-803 cells (99). CCNG2 is a regulator of cell proliferation and induces G1/S phase cell cycle arrest (100,101). CCNG2 expression is decreased in GC (102). Based on data derived from TCGA, miR-240 is over-expressed in GC tissues in comparison to matching normal tissues (Figure 2).
Other miRs Affecting Growth of Gastric Cancer
miR-10b. miE-10b (Figure 5B) promotes proliferation, invasion, colony formation and EMT of HGC27 GC cells (103). MKN74 cells overexpressing miR-10b show increased tumor volume and weight and metastasis to the liver after subcutaneous implantation in nude mice (103). CUB and sushi multple domains 1 (CSMD1) was identified as a target of miR-10b (103). Expression of CSMD1 was negatively correlated with expression of miR-10b in GC tissues (103). Inhibition of CSMD1 leads to activation of the NFĸB pathway resulting in up-regulation of cyclin D1 (CCND1) and EMT markers (103). CSMD1 is a single membrane spanning protein which contains 14 N-terminal CUB domains that are separated from each other by a SUSHI domain followed by an additional 15 tandem SUSHI domain segment and functions as a complement inhibitor (104). In melanoma cells, CSMD1 exhibits anti-tumor activity by inhibiting cell-cycle progression and apoptosis via the SMAD pathway (105). Loss of CSMD1 has been observed in head-and-neck squamous cell carcinoma, lung- and breast cancer (106).
miR-130a. miR-130a (Figure 5B) is elevated in GC cell lines (107). In BGC823 cells, miR-130a promotes proliferation and inhibits cell-cycle arrest in G1 phase, colony formation, cell invasion and migration as well as cell adhesin by targeting collapse response mediator protein 4 (CRMP4) (107). CRMP4 reversed the in vitro effects of miR-130a as described above (107). miR-130a functions as an oncomir. miR-130a accelerates TG of BGC823 cells in nude mice and depletion of CRMP4 with small hairpin RNA (shRNA) results in retardation of TG (107). CRMPs are a family of five cytosolic proteins which are expressed in the nervous system during development and through interactions with microtubules play important roles in axon formation and growth cone guidance and collapse (108). They are implicated in proliferation, apoptosis, differentiation, progression and metastasis of tumors (109). In melanoma cells, CRMP1 plays an anti-tumoral role in cell-cycle regulation and controlling apoptosis via the SMAD pathway (110). It was shown that VEGF promotes GC development by up-regulation of CRMP4 (111).
miR-135b-5p. miR-135b-5p (Figure 5B) expression in GC tissues is significantly higher compared to normal tissues (112). SGC-7901 GC cells infected with a lentivurs expressing a miR-135b-5p inhibitor exhibit reduced proliferation, suppressed cell-cycle progression and invasiveness and promotion of apoptosis (112). In vivo, this cell line has characteristics of reduced tumor growth in nude mice (112). Chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containg family member 3 (CMTM3) was identified as a direct target of miR-135b-5p (112). CMTM3 is member of a family of nine genes in humans of the chemokine-like factor gene family similar to the chemokine and transmembrane 4 superfamilies of signaling molecules (113). It was previously shown that CMTM3 inhibits GC cell growth through apoptosis and its knockdown promotes metastasis of GC cells via the STAT3/Twist1/EMT signaling pathway (114-116). CMTM3 also decreases epidermal growth factor receptor (EGFR) expression and EGFR-mediated tumorigenicity in GC (117). Altogether, CMTM3 functions as a TS in GC and CMTMs excert TS functions in additional types of cancers (118).
miR-448. miR-448 (Figure 5B) is over-expressed in GC and is associated with poor survival (119). miR-448 promotes growth and carcinogenicity of MKN87 and SGC7901 GC cells in vitro and in vivo (119). Lysine (K)-specific demethylase 2B (KDM2B) was identified as a direct target of miR-448. Suppression of KDM2B promotes glycolysis in GC cells and miR-448 is a positive regulator of glycolysis (119). Inhibition of of KDM2B induces expression of transcription factor MYC and glycolysis (119). KDM2B removes methyl groups from H3K36me2 and H3K4me3 of histones and inhibits glycolysis of cancer cells which are addicted to glycolysis in contrast to differentiated cells which use mitochondrial oxidative phosphorylation for energy production (119). MYC is known as inducer of glucose uptake and lactate production in cancer cells (120). However, from a general point of view, one should keep in mind that KDM2B is a double-edged sword in the regulation of cancer development which can act in a context-dependent manner as a TS or as an oncogene (121).
miR-487a. miR-487a (Figure 5B) promotes proliferation and suppresses apoptosis of GC in vitro and knockdown of miR-487a has opposite effects (122). miR-487a enhances growth of GC xenografts (122). T cell intracellular antigen-1 (TIA-1) has been identified as a direct target of miR-487a (122). TIA-1 contains three RNA recognition domains, the latter two have been shown to be important for RNA binding and selectivity (123). TIA-1 alters both co-transcriptional and post-transcriptional RNA processing and binds to VEGF-A mRNA (124). Alternative splicing of VEGF induced by TIA-1 influences the angiogenic capability of colorectal cancer (125). TIA-1 also acts as a stress-induced translational inhibitor localizing to stress granules containg polyA RNA (126). Alternative splicing, export and translational regulation have been shown to contribute to tumor formation and progression and these events are attractive targets for therapeutic intervention (127,128).
Technical Aspects
Oncomirs can be inhibited at several stages of their generation: at the level of transcription by RNA Pol II or III, at the level of precursor cleavage by DICER and at the level of functional inhibition of mature RNA, resulting in mRNA degradation and functional repression (129). The latter is the most popular mode of intervention. miRs inhibit several mRNAs, wheras siRNA inhibits only one specific mRNA (130). siRNAs are 21-23 nt RNA duplexes with two nts 3’-overhang fully complementary to mRNA, whereas miRs are 19-25 nts RNA duplexes with two nts 3’-overhang with complementary binding in the seed region (nts 2 to 7 at the 5’-end) of the corresponding miR to the 3’-UTR of the mRNA (130). siRNA mediates endonucleolytic cleavage of the corresponding mRNA, wheras miRs preferentially induce degradation of mRNAs and translational repression (129,130).
Anti-miR-oligonucleotides (AMO) are designed to bind a sequence complimentary to mature miRs. Incorporation of a phosphoro-thioate backbone and ribose 2’-OH modification (2’-F, 2’-O-methyl, 2’-O-methoxyethyl) have improved binding and pharmaco-kinetic and pharmako-dynamic properties of AMO’s (130). Anti-miRs with a 2’-O-methoxyethyl modification are referred to as antagomirs. Also, incorporation of locked nucleic acids (LNA) which contain an additional bridge between the 2’-oxygen and the 4’-carbon of the pentose have significantly improved the drug-like properties of AMO’s (131).
Peptide nucleic acids (PNA) are under evaluation as miR-inhibitory agents (132). In these agents the deoxy-phosphate backbone of nucleic acids is replaced by a polyamide chain of N-aminoethyl-glycyl units to result in polymers with an artificial backbone structure as a main chain with similarity to DNA or RNA (132). PNAs are resistant to nucleases, have stronger affinity and greater specificity for DNA and RNA than natural nucleic acids (132).
Identification of small molecule inhibitors of miRs is under active investigation. Several inhibitors which inhibit transcription of defined miRs have been identified, however specificity issues and target deconvolution of the identified compounds have to be tackled (133). Identification of compounds which target secondary elements in human miR hairpin precursors have been reported (134). It has been shown that a secondary structure of pre-miRs contains a narrow groove to which a positively charged compound can bind at nM affinity, making it a druggable candidate (135). Combination of bioinformatics and high throughput screening has revealed compounds which inhibit DICER cleavage sites in pre-miRs (136).
miR sponges are DNA constructs which contain artificially designed, tandemly reiterated miR-binding sites thus competing for miRs with the mRNA under consideration (137). They have to be transduced into the corresponding recipient cells (137). Furthermore, oncomirs have been knocked-out by making use of the genome-editing tool clustered regularly interspaced short palindromic repeats- CRISPR associated (CRISPR-CAS) (138,139).
However, many additional issues have to be optimized case-by- case for optimization of therapeutic efficacy of miR-inhibitors. Extended circulation time by PEGylation [attachment of polyethylene glycol (PEG)] polymer chains attached to miRs has been achieved (140). Optimization of delivery is a crucial process for achieving therapeutic efficacy. Cationic complexes which interact with negatively charged RNA through electrostatic interactions have been created with synthetic polyethyleneimine (PEI) as early generation polymers for RNA delivery (141). Also, dendrimers, branched synthetic polymers are under evaluation as delivery agents (142,143). Polymers also promote endosomal escape of miRs thus avoiding endosomal-lysosomal RNA degradation. Poly (lactic-co-glycolic acid) (PLGA) is an FDA-approved synthetic biodegradable polymer for delivery of miR-inhibitors (144). Moreover, the use of cyclodextrins as delivery agents has been explored (145). Another class of delivery agents are lipid-based agents. These agents are composed of cationic lipids and liposomes which can form complexes with RNA through electrostatic interactions (146). Also, lipolyplexes composed of polymers and lipids are under investigation as delivery agents (147). Conjugation of miRs to specific ligands such as N-acetylgalactosamine, a triantennary N-acetylgalactosamine, high-affinity ligand specific for the asialoglycoprotein receptor, enhances potency of anti-sense oligonucleotides 6-10-fold in mouse liver (148). Several ciritical issues are not discussed in further details in this review such as toxicity issues, cytokine release syndrome, delivery into target tissues, efficient release of miR-related agents from endosomes, optimization of pharmaco-kinetic and pharmaco-dynamic properties, excretion by the kidneys and unspecific hybridization (149-155).
Synopsis. We have identified 31 miRs covering 26 different targets which promote growth of GC cells in vitro and in vivo as xenografts. Five miRs (miRs -10b, 151-5p, -187, 532-3p and -589) additionally have an impact on metastasis. Thirteen of the identified miRs (-19b, -20s, -25, -92a, -106a, -135a, -187, -221-3p, -340a, -421, -493, -575 and -589 correlate with worse prognosis in GC patients. The oncomirs identified are up-regulated in GC tissues, the corresponding targets are down-regulated. The homogeneity of expression of the target candidates miRs should be investigated in more details. As outlined previously GC patients have a high relapse rate after surgical resection and following chemotherapy. It would be of interest to investigate, whether inhibition of any of the identified miRs has an impact on established metastases in preclinical in vivo models.
miRs Landscape Cancer
miR-related therapeutics may be particulary suited for complex, multigenic disorders such as cancer, since they can interfere with several pathways. The first micro-RNA based therapeutic evaluated in clinical studies in cancer patients was MRX-34 by Mirna Therapeutics (156,157). MRX-34 is an intra-venously injected liposomal formulation of a miR-34 mimetic. miR-34 acts as a TS and inhibits proliferation, invasion by inhibition of WNT, NOTCH, TGFβ and EMT-related transcription factors (156,157). Repression of miR-34 target genes such as forkhead box protein P1 (FOXP1), B-cell lymphoma 2 (BCL2), histone deacetylase 1 (HDAC1) and cyclin B1 (CTNNB1) was demonstrated in cancer patients, however, clinical studies were halted due to multiple immune-related side effects (156,157). Another agent is a miR-16-based mimetic which is administered as EGFR-conjugated bacterial mini-cells (158). The agent was intra-venously injected into recurrent patients with malignant pleural mesothelioma with signs of radiologic progression after chemotherapy (NCI02369198). Twenty-two patients were enrolled and one objective response (5%), in 15 patients (68%) stabilisation of disease and in 6 patients (27%) progression of disease was observed with an acceptable safety profile (158). miR-16 preferentially targets BCL2 (159). Inhibition of miR-155 is pursued by miRagen. miR-155 acts oncogenically in many haematological diseases and its expression correlates with prognosis in lymphoma and leukemic diseases (160). Cobomersen is a 14 nucleotides mixed-type (deoxynucleotide plus LNA phosphorothioate-based oligonucleotide which inhibits proliferation of tumor cells and T-cell activation and simultaneously regulates JAK/STAT, MAPK/ERK and PI3K/AKT signalling (160). In 68 patients with cutaneous T-cell lymphoma (CTCL), acute T-cell leukemia/lymphoma (ATLL), diffuse B-cell lymphoma (DBCL) and chronic lymphpcytic leukemia (CLL) cobomersen was administered for 2 years, reversion of the disease genetic signature and no serious side effects were observed (www.miragen.com). In CTLL patients cobomersen has reduced lesion severity after i.v. injection in a Phase I study. It remains to be seen whether miR-based therapeutic agents have a bright future in the space of cancer therapy.
Authors’ Contributions
AN and UHW: design and writing of the paper; FB: bio-informatic analysis.
References
Articles from Cancer Genomics & Proteomics are provided here courtesy of International Institute of Anticancer Research
Full text links
Read article at publisher's site: https://doi.org/10.21873/cgp.20237
Read article for free, from open access legal sources, via Unpaywall:
https://cgp.iiarjournals.org/content/cgp/18/1/1.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.21873/cgp.20237
Article citations
miRNA on the Battlefield of Cancer: Significance in Cancer Stem Cells, WNT Pathway, and Treatment.
Cancers (Basel), 16(5):957, 27 Feb 2024
Cited by: 0 articles | PMID: 38473318 | PMCID: PMC10931375
Review Free full text in Europe PMC
Salidroside inhibited the proliferation of gastric cancer cells through up-regulating tumor suppressor miR-1343-3p and down-regulating MAP3K6/MMP24 signal molecules.
Cancer Biol Ther, 25(1):2322206, 04 Mar 2024
Cited by: 2 articles | PMID: 38436092 | PMCID: PMC10913707
The seen and the unseen: Molecular classification and image based-analysis of gastrointestinal cancers.
Comput Struct Biotechnol J, 20:5065-5075, 12 Sep 2022
Cited by: 1 article | PMID: 36187924 | PMCID: PMC9489806
Review Free full text in Europe PMC
Glaucocalyxin A Inhibits the Malignancies of Gastric Cancer Cells by Downregulating MDM2 and RNF6 via MiR-3658 and the SMG1-UPF mRNA Decay Pathway.
Front Oncol, 12:871169, 22 Jun 2022
Cited by: 2 articles | PMID: 35814430 | PMCID: PMC9258495
MicroRNAs and Corresponding Targets in Esophageal Cancer as Shown In Vitro and In Vivo in Preclinical Models.
Cancer Genomics Proteomics, 19(2):113-129, 01 Mar 2022
Cited by: 1 article | PMID: 35181582 | PMCID: PMC8865044
Review Free full text in Europe PMC
Go to all (9) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
In vitro and in vivo effects of miRNA-19b/20a/92a on gastric cancer stem cells and the related mechanism.
Int J Med Sci, 15(1):86-94, 01 Jan 2018
Cited by: 17 articles | PMID: 29333091 | PMCID: PMC5765743
MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2.
Oncotarget, 7(32):51943-51954, 01 Aug 2016
Cited by: 84 articles | PMID: 27409164 | PMCID: PMC5239526
MiR-19b/20a/92a regulates the self-renewal and proliferation of gastric cancer stem cells.
J Cell Sci, 126(pt 18):4220-4229, 18 Jul 2013
Cited by: 81 articles | PMID: 23868977
Down-regulated MicroRNAs in Gastric Carcinoma May Be Targets for Therapeutic Intervention and Replacement Therapy.
Anticancer Res, 41(9):4185-4202, 01 Sep 2021
Cited by: 7 articles | PMID: 34475038
Review




