Abstract
Free full text

How the PhoP/PhoQ System Controls Virulence and Mg2+ Homeostasis: Lessons in Signal Transduction, Pathogenesis, Physiology, and Evolution
SUMMARY
The PhoP/PhoQ two-component system governs virulence, Mg2+ homeostasis, and resistance to a variety of antimicrobial agents, including acidic pH and cationic antimicrobial peptides, in several Gram-negative bacterial species. Best understood in Salmonella enterica serovar Typhimurium, the PhoP/PhoQ system consists o-regulated gene products alter PhoP-P amounts, even under constant inducing conditions. PhoP-P controls the abundance of hundreds of proteins both directly, by having transcriptional effects on the corresponding genes, and indirectly, by modifying the abundance, activity, or stability of other transcription factors, regulatory RNAs, protease regulators, and metabolites. The investigation of PhoP/PhoQ has uncovered novel forms of signal transduction and the physiological consequences of regulon evolution.
INTRODUCTION
The PhoP/PhoQ two-component system consists of the DNA-binding protein PhoP and the sensor PhoQ and is found in a number of gammaproteobacterial species. The PhoQ protein responds to cytoplasmic and extracytoplasmic signals by modifying the phosphorylation status of PhoP: when the bacterium experiences activating signals, PhoQ autophosphorylates from adenosine triphosphate (ATP) at a conserved histidine residue. The phosphate in phosphorylated PhoQ is then captured by PhoP (1,–3) at a conserved aspartate residue within its N-terminal domain. In addition to its autokinase and phosphotransferase activities, PhoQ exhibits phosphatase activity towards phosphorylated PhoP (PhoP-P) (1,–4). In vitro, both PhoP-P and PhoP bind to specific DNAs (5,–13). In vivo, however, only PhoP-P binds to its target DNAs when it is present in physiological amounts (14). Upon binding to DNA, PhoP-P increases transcription of PhoP-activated genes and reduces the transcription of PhoP-repressed genes (12,–16). These PhoP-regulated transcriptional changes result in a variety of phenotypic modifications that enable bacteria to resist killing by antimicrobial agents and to cope with nutritional stresses (17,–23) (Fig. 1).
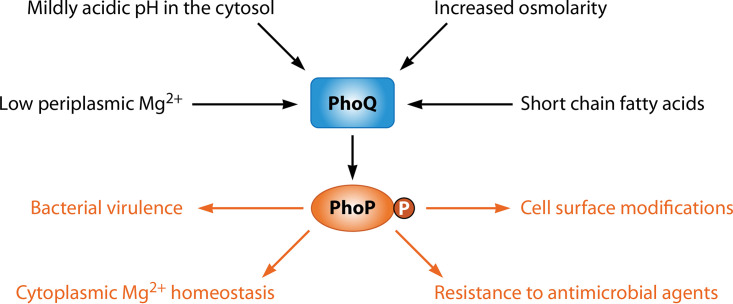
Regulation of and by the PhoP/PhoQ two-component system. The sensor PhoQ is activated by the signals indicated in black. Upon phosphorylation by PhoQ, the regulator PhoP controls the activities indicated in orange.
The phoP gene was originally identified during a genetic search for Salmonella enterica serovar Typhimurium (S. Typhimurium) mutants defective in a nonspecific acid phosphatase (24). Two classes of mutants were recovered: those with mutations mapping to phoN, which was proposed to be the structural gene for a nonspecific acid phosphatase, and those mapping to phoP, which was proposed to be a regulatory gene. We now know that phoP controls expression of the nonspecific acid phosphatase gene phoN (13, 25, 26). The phoP gene exhibits a broad phylogenetic distribution within enteric bacteria and is also found in Gram-negative species outside the family Enterobacteriaceae (27, 28) (Table 1). By contrast, phoN is a horizontally acquired gene in S. Typhimurium and, as such, exhibits a limited, sporadic presence in other enteric bacteria (29). The regulatory relationship between the phoP and phoN genes established in S. Typhimurium (13, 25, 26) has not been reported in other bacterial species. Nonetheless, PhoP controls physiological pathways that intersect with phosphate metabolism in organisms lacking the phoN gene (30).
TABLE 1
Homologous genes to the enterobacterial phoP/phoQ system in other bacterial species
| Genus | Genes | Identity (%)a | Experimentally demonstrated functions | Notes | Key reference(s) |
|---|---|---|---|---|---|
| Salmonella | phoP/phoQ | 100 | Mg2+ homeostasis, antimicrobial peptide resistance, growth in acid pH, virulence | 18, 68 | |
| Shigella | phoP/phoQ | 94 | Antimicrobial peptide resistance, virulence | 242 | |
| Escherichia | phoP/phoQ | 93 | Mg2+ homeostasis, antimicrobial peptide resistance, growth in acid pH, PMF generation, virulence | 362 | |
| Serratia | phoP/phoQ | 83 | Mg2+ homeostasis, antimicrobial peptide resistance, growth in acid pH, virulence | 403 | |
| Sodalis | phoP/phoQ | 82 | Antimicrobial peptide resistance | More functions in free-living Sodalis (e.g., Mg2+ homeostasis) | 401, 402 |
| Yersinia | phoP/phoQ | 80 | Mg2+ homeostasis, growth in acid pH, oxidative/osmotic stress resistance, survival in macrophages | Only minor virulence defects of a phoP mutant | 244 |
| Dickeya | phoP/phoQ | 80 | Mg2+ homeostasis, antimicrobial peptide resistance, growth on acid pH, pectin metabolism, apoplast alkalinization, virulence | 392, 393 | |
| Edwardsiella | phoP/phoQ | 79 | Mg2+ homeostasis, temperature sensing, virulence | 406 | |
| Pectobacterium | pehR/pehS or phoP/phoQ | 78 | Pectin metabolism, virulence | 409 | |
| Pseudomonas | phoP/phoQ | 54 | Mg2+ homeostasis, antimicrobial peptide resistance | PhoQ may not be able to sense AMPs; the phoQ mutant is more resistant to AMPs | 86, 414 |
The phoP gene was rediscovered when three research groups reported that S. Typhimurium phoP- and phoQ-null mutants are highly attenuated for virulence, exhibiting a median lethal dose five orders of magnitude higher than that of wild-type S. Typhimurium in mice inoculated via the peritoneal route (17, 27, 31, 32). Both phoP and phoQ mutants are defective for survival inside mammalian macrophages and hypersusceptible to killing by several antimicrobial peptides, including the rabbit-derived defensin NP1 and the frog-derived magainin 2 (19, 27). Curiously, the pho-24 allele, a missense mutation in phoQ specifying a PhoQ protein with the T48I substitution that causes high expression of PhoP-activated genes, attenuates S. Typhimurium virulence as much as inactivation of the phoP or phoQ genes (18, 33) (Fig. 2A). (Please note that in some of the early literature, S. Typhimurium strains with the pho-24 allele are referred to as PhoPc for PhoP constitutive even though the pho-24 allele corresponds to a mutation in phoQ, not phoP.) Given that the phoPQ operon specifies a two-component regulatory system, these results established that S. Typhimurium virulence is a regulated process (as opposed to a behavior always on display) and that the PhoP/PhoQ system plays a dominant role in S. Typhimurium pathogenicity. In addition, these results suggested that the bacterial ability to resist killing by antimicrobial peptides is necessary for virulence, a notion supported by a subsequent study (19).
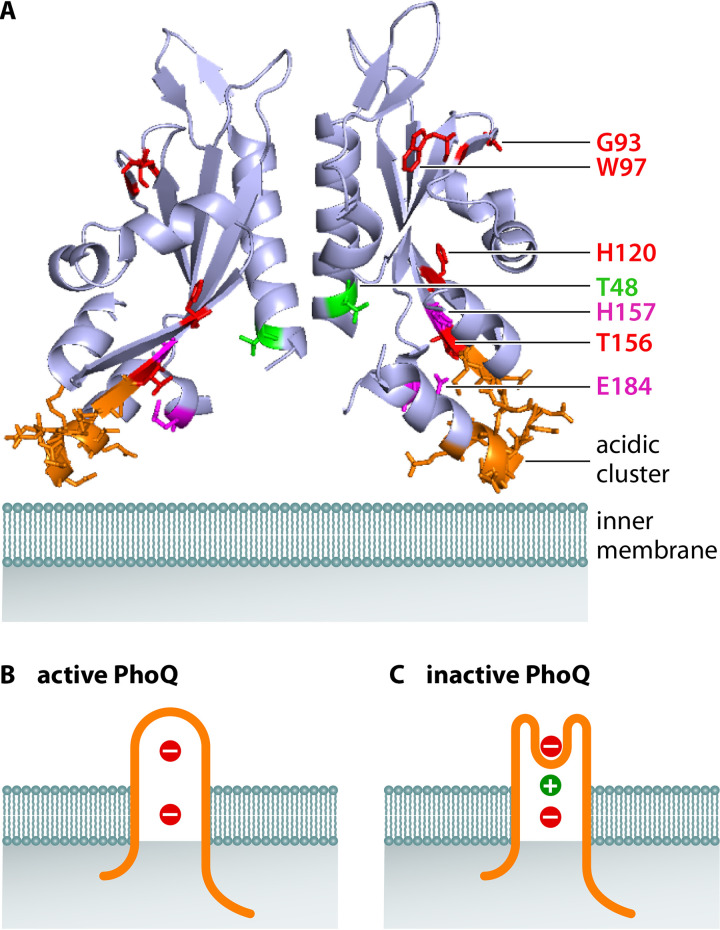
Structure of PhoQ’s periplasmic domain and model of PhoQ sensing of divalent cations. (A) X-ray structure of the periplasmic domain of the E. coli PhoQ dimer (PDB 3BQ8) (64). The expected position of the inner membrane is in light teal; residues in red correspond to those in which there is direct evidence of a loss of cation binding upon substitution (60); residues in purple correspond to those in which there is indirect evidence in cation sensing through downstream effects on a PhoP-activated gene (62); the residue in green is involved in sensing Ca2+ but not Mg2+ (61); acidic cluster in orange has a minor impact on cation sensing despite being often discussed in the literature (62). (B) In the absence of divalent cations, the membrane negative charges repel the negatively charged periplasmic domain of PhoQ, putting the lever in the “up” position and promoting PhoQ autophosphorylation. PhoQ is in orange, divalent cations in green, and negative charges in the inner membrane in red. (C) When present, the positively charged divalent cations bridge the negative charges in PhoQ and the inner membrane, bringing the lever to the “down” position and hindering PhoQ autophosphorylation.
The presence of phoP and phoQ in nonpathogenic species (27, 28, 34), such as commensal Escherichia coli, indicates that, in addition to controlling virulence, the PhoP/PhoQ system regulates physiological processes common to both pathogens and nonpathogens. One such process is homeostasis for Mg2+, the most abundant divalent cation in all living cells (35). The PhoP/PhoQ system responds to changes in the Mg2+ concentration in its surroundings by altering expression of different classes of genes playing a role in Mg2+ homeostasis: genes responsible for chemical modifications of the bacterial cell surface at sites normally neutralized by Mg2+ and other divalent cations (36); genes responsible for importing Mg2+ from the periplasm into the cytoplasm (35); and genes furthering protein homeostasis that, among various roles, enable protein synthesis to continue and ATP-dependent proteolysis of functional proteins to decrease (37) when the cytoplasmic Mg2+ concentration drops below the threshold that compromises ribosome assembly and activity (38).
The investigation of the PhoP/PhoQ system led to the discovery of the first example of a signal transduction system that responds to Mg2+ as the primary signal (18); the first example of a physiological connection among different two-component regulatory systems (39,–42); the demonstration that the chemical form of the lipopolysaccharide (LPS) is dynamic, changing in response to environmental conditions, such as those S. Typhimurium experiences inside a mammalian host (43, 44); the connection among phosphate limitation, Mg2+ starvation, and protein synthesis (30); and how bacterial regulons evolve (28, 45, 46). In addition, the PhoP/PhoQ system has been used to investigate the evolution of interacting proteins (47).
The PhoP/PhoQ system from S. Typhimurium and other Gram-negative bacteria is not the physiological equivalent of the PhoP/PhoR system of Gram-positive bacteria (48, 49), even though both PhoP/PhoQ and PhoP/PhoR are two-component regulatory systems (50) (Table 2). PhoP/PhoQ is activated in low Mg2+ and promotes expression of genes mediating bacterial survival in low-Mg2+ environments (18), whereas PhoP/PhoR is activated in bacteria experiencing low-phosphate conditions and promotes expression of genes that help bacteria cope with low-phosphate environments (48). The physiological role played by PhoP/PhoR in Gram-positive bacteria resembles that of the PhoB/PhoR system of S. Typhimurium and other enteric bacteria (51). The PhoB/PhoR system is activated not only in response to low environmental phosphate but also in low cytoplasmic Mg2+ (30) (discussed below under “The PhoP-Dependent Connection among Low Cytoplasmic Mg2+, Protein Synthesis, and the Phosphate-Responding PhoB/PhoR System”). In addition, the enteric PhoP protein is not the functional sequelog of the PhoP protein from Mycobacterium tuberculosis (52) despite the corresponding genes being required for virulence in their respective species.
TABLE 2
Ambiguously named genes which may be mistaken for homologs to the enterobacterial phoP/phoQ system
| Genus | Genesb | Identity (%)a | Functions | Notes | Key reference |
|---|---|---|---|---|---|
| Saccharopolyspora | phoP/phoR (SACE_6965/SACE_6966)* | 34 | Phosphate and nitrogen metabolism | 240 | |
| Streptomyces | phoP/phoR (SCO4230/SCO4229)* | 33 | Phosphate and nitrogen metabolism, antibiotic synthesis | 49 | |
| Mycobacterium | phoP/phoR | 32 | Virulence, Mg2+ homeostasis, lipid biosymthesis; resistance to some oxidative compounds and antibiotics | Likely homologous to the B. subtilis phoP/phoR (41% identity) | 400 |
| Bacillus | phoP/phoR | 31 | Phosphate metabolism | 415 |
Here, we discuss the signals activating the sensor PhoQ, the DNA sequences recognized by PhoP, the genes regulated by PhoP, the cellular activities controlled by PhoP/PhoQ, the feedback mechanisms that alter the amount of active PhoP protein over time, and the physiological role that the PhoP/PhoQ system plays in different bacterial species. The discussion is based primarily on the S. Typhimurium PhoP/PhoQ system because S. Typhimurium is the organism in which the majority of the research on PhoP/PhoQ has been reported. Critically, many of the lessons learned during these investigations apply not only to the PhoP/PhoQ system of other species but also to other regulatory systems and physiological interactions even when the specific gene product carrying out a given activity differs across species. In addition, we describe the similarities and differences that the PhoP/PhoQ system exhibits across bacterial species in terms of signal input, transcriptional output, and interactions with other regulatory systems.
WHAT THE SENSOR PhoQ SENSES: LOOKING INSIDE AND OUTSIDE THE CYTOPLASM FOR SIGNALS
The S. Typhimurium PhoQ protein harbors two transmembrane domains that define a 146 amino acid long periplasmic region and two cytoplasmic domains: a 45 amino acid long domain at the very N terminus and a C-terminal 259 amino acid long domain. The latter domain harbors the region of sequence similarity shared with other sensors of the two-component system family that is responsible for dimerization, autophosphorylation, and interaction with the cognate regulator (50).
Here, we consider the signals detected by the PhoQ protein and the physiological significance of such signals in terms of the genes controlled by PhoP and the phenotypes displayed by phoP and phoQ null mutants. This analysis provides a critical understanding of the environments that alter PhoP-P abundance because the wild-type PhoP protein is phosphorylated neither from sensors other than PhoQ nor from acetyl phosphate (53), a small molecular weight phosphate donor used by some regulators of the two-component system family to autophosphorylate (54). By promoting PhoP dimerization, phosphorylation enables PhoP to bind target DNA sequences and to modify gene expression in vivo (14). Therefore, nonphysiological dimerization is likely responsible for the reported transcription of PhoP-activated genes taking place in the absence of phoQ or with nonphosphorylatable PhoP variants when the PhoP protein is expressed at 13 times the levels achieved under high inducing conditions in wild-type S. Typhimurium (55).
Mg2+ and Other Divalent Cations
PhoQ senses periplasmic Mg2+, providing the first example of a biological sensor that responds to Mg2+ as primary signal (18). When S. Typhimurium experiences >2 mM Mg2+ in its surroundings, PhoP-activated genes are fully repressed, and PhoP-repressed genes are derepressed (18, 56). As the Mg2+ concentration in the growth medium decreases, the level of PhoP-P increases, resulting in transcription of PhoP-activated genes and repression of PhoP-repressed genes (15). Some PhoP-activated genes are transcribed during growth at 1 mM Mg2+ (56), whereas others require prolonged growth in 10 μM Mg2+ to activate the various feedback loops that increase the amount of PhoP-P to the levels required to transcribe those genes (57, 58). The phoP-activated gene mgrR of E. coli is unusual in that its expression is phoQ independent and reaches 50% of the maximum levels during growth in 1 mM Mg2+ (59). As discussed in the section “PhoP-Activated Regulatory RNAs,” the mgrR expression behavior is ascribed to the properties of its promoter (59).
That PhoQ activity increases in response to low extracytoplasmic Mg2+ makes physiological sense given the biochemical activities of some PhoP-regulated gene products (35) and given that both phoP and phoQ mutants are defective for growth in low Mg2+ (18). For example, divalent cations, primarily Mg2+, neutralize the majority of the negative charge conferred by the phosphate residues in the LPS, which occupies the outer leaflet of the outer membrane in Gram-negative bacteria. Therefore, when bacteria experience low Mg2+ in their surroundings, PhoP-activated gene products covalently modify the LPS (36). The LPS modifications help bacteria in two ways: first, they avoid electrostatic repulsion between phosphate residues in adjacent LPS molecules; and second, they free Mg2+ so that it can be imported from the periplasm and into the cytoplasm by Mg2+ transporters transcriptionally activated by PhoP. The imported Mg2+ helps satisfy the plethora of cytoplasmic factors that exhibit a strict dependence on Mg2+.
When sensed by the periplasmic domain of PhoQ, Mg2+ represses PhoQ’s autokinase activity (60) while promoting its phosphatase activity (4). These two activities decrease PhoP-P amounts both by reducing the amount of phosphorylated PhoQ available to serve as phosphodonor to PhoP and by stimulating dephosphorylation of PhoP-P.
PhoQ’s periplasmic domain is necessary for the response to Mg2+. This is because a chimeric protein in which the 190 N-terminal amino acids of PhoQ, including the full periplasmic domain, are replaced by the equivalent region of the two-component sensor EnvZ is no longer responsive to changes in the Mg2+ concentration in the bacterium’s surroundings, resulting in constitutive high expression of PhoP-activated genes in both high and low Mg2+ (18). In addition, the fluorescence of the single tryptophan residue in the purified periplasmic domain of the S. Typhimurium PhoQ protein changes in the presence of Mg2+ (61) (Fig. 2A). Moreover, mutation of the periplasmic residues G93, W97, H120, and T156 decreases the Mg2+ sensitivity of the full-length PhoQ present in its normal inner membrane location (60) (Fig. 2A). These four residues are conserved in the PhoQ proteins of other gammaproteobacterial species, including several with Mg2+-responding PhoP/PhoQ systems (60).
A cluster of acidic amino acids (residues145 to 154 in the S. Typhimurium PhoQ protein) has been proposed to participate in Mg2+ sensing (62) (Fig. 2A). Likewise, H157, M155, and E184 have also been implicated in Mg2+ sensing because strains expressing PhoQ variants in which these residues were substituted exhibited increased activity of the PhoP-activated PhoN protein in high Mg2+ (62) (Fig. 2A). However, the cluster of acidic amino acids is not conserved in the PhoQ proteins from other enteric species that silence PhoP in response to divalent cations; how these PhoQ variants interact with Mg2+ is currently unknown.
Ca2+ and Mn2+ also act on PhoQ, resulting in PhoP inactivation (18); by contrast, Ni2+, Cu2+, and Ba2+ had no effect when tested at concentrations up to 300 μM (note that half-maximal repression of PhoP-activated genes was achieved at 50 μM for Ca2+ and 0.85 μM for Mg2+) (18). The Mg2+ and Ca2+ binding sites in PhoQ appear to be distinct because the effect of the two cations is cumulative and also because the T48I substitution in PhoQ’s periplasmic region abolishes sensing of Ca2+ but not of Mg2+ in vivo (18, 61) (Fig. 2A). However, the NMR spectra of the purified periplasmic domain of PhoQ bound to Mg2+ and Ca2+ are similar (62), implying that both cations promote similar conformational changes in PhoQ’s periplasmic region. Thus, the nuclear magnetic resonance (NMR) spectra of variants of PhoQ’s periplasmic region in the presence of different divalent cations in vitro do not readily reflect the behavior of the full-length PhoQ protein in vivo.
An X-ray structural analysis of a Ca2+-bound periplasmic PhoQ domain from S. Typhimurium led to the proposal that periplasmic divalent cations regulate PhoQ activity by a lever mechanism (62). According to this proposal, cations anchor PhoQ to the membrane by bridging PhoQ’s nonconserved acidic patch residues to the negative charges provided by the phospholipid head groups in the inner membrane. In the absence of divalent cations, electrostatic repulsion between PhoQ’s acidic patch and the membrane phospholipids is proposed to result in PhoQ autophosphorylation (62, 63) (Fig. 2B). However, the structure of the Ca2+-bound periplasmic PhoQ domain appears to be a tetramer (PDB 1YAX) (62), which is puzzling because the expected position of the transmembrane domains makes it impossible for such a structure to exist in vivo (i.e., PhoQ is a dimer that harbors two transmembrane domains per monomer). In addition, several Ca2+ atoms seem to interact with the acidic clusters on both dimers, hence artificially bridging them, which might have contributed to the observed structural differences between the free and Ca2+-bound structures reported in the NMR studies (62).
When placed onto the X-ray structure of the periplasmic domain of the E. coli PhoQ protein (PDB 3BQ8) (64), the amino acids involved in cation sensing are clustered near the transmembrane domains (Fig. 2B). These amino acids include T48, a substitution of which by isoleucine alters the response to Ca2+ in vivo (18, 61) (Fig. 2A), supporting the lever model of PhoQ activation (Fig. 2B and andC).C). This model, however, does not account for the role of G93 and W97 in Mg2+ sensing (60) (Fig. 2A), as these residues are situated opposite the transmembrane domain in both structures. Therefore, the current model will have to be refined to account for these two residues and for the unique role that T48 plays in the response to Ca2+.
Antimicrobial Peptides
In the laboratory, PhoQ is activated by sublethal concentrations of different cationic antimicrobial peptides (CAMPs) (65). The activating peptides include the alpha-helical C18G, a synthetic derivative of the C terminus of the human platelet factor IV with improved antimicrobial activity (66), the beta-sheet peptide protegrin-1, and polymyxin B, a cyclic peptide with a fatty acid tail from a soil bacterium (65). By permeabilizing the outer membrane (65), these peptides have the potential to reach PhoQ’s periplasmic domain.
PhoP/PhoQ activation by CAMPs makes intuitive sense given that phoP and phoQ mutants are highly susceptible to killing by certain CAMPs (17, 19, 21, 67) and that S. Typhimurium faces CAMPs in different tissues during infection of mammalian hosts. Moreover, incubation with sublethal concentrations of C18G increased resistance not only toward C18G but also toward protegrin-1 and polymyxin B (65). However, the strength of PhoQ activation and of peptide antimicrobial activity against phoP mutant bacteria are not correlated (68). That is, the human-derived peptide LL37 induces expression of the PhoP-activated gene pagP but does not preferentially kill a phoP mutant relative to wild-type S. Typhimurium (68). In the case of derivatives of the peptide bactenecin, there is an inverse correlation between the ability to promote transcription of a PhoP-activated gene and antimicrobial potency (69). PhoQ activation is actually correlated with peptide hydrophobicity and amphipathicity. The Shai laboratory found that bacterial preincubation with several natural and artificial CAMPs at subinhibitory concentrations failed to induce CAMP resistance (68). This is in contrast to the reports by the Miller group for some CAMPs (65) and the reports of others for a wide range of antimicrobial compounds (summarized in reference 70). In sum, there is no strict one-to-one correlation between PhoQ activation by a CAMP and preferential toxicity toward phoP and phoQ mutants by that CAMP.
Depending on the nature of the CAMP, PhoQ activation is proposed to be either a competitive inhibition of cation binding (71) (Fig. 2B and andC)C) or a sequestration of cations in the medium by specific histidine residues (68). In agreement with the former notion, an excess of Mg2+ in the medium limits PhoQ activation by a CAMP (71). In addition, residues T156 and E184, both involved in Mg2+ sensing, contribute to CAMP sensing (71).
When a variety of natural and synthetic CAMPs were investigated, PhoQ was found to efficiently respond to peptides with a high positive charge and high hydrophobicity (68). This observation is coherent with the lever model discussed in the previous section because amphipathic, positively charged CAMPs would insert between PhoQ and the membrane and also because a CAMP’s large size, compared to Mg2+, would hold the lever in the “up” (i.e., active) position (Fig. 2B and andCC).
The murine cathelicidin-related antimicrobial peptide (CRAMP) induces expression of PhoP-activated genes in laboratory media but not when S. Typhimurium is inside murine macrophages (72). Although the phoP-null mutant survives less well than wild-type S. Typhimurium in both CRAMP-deficient and -proficient macrophages (73), the defect of the phoP mutant is more pronounced in CRAMP-proficient macrophages (73), in agreement with the phoP-null mutant being hypersensitive to killing by CRAMP in vitro (74). The human equivalent of CRAMP (i.e., LL37) is not induced in human monocyte-derived macrophages and does not appear to play a major role in bacterial killing (75). These studies indicate that the PhoP/PhoQ system confers protection from CRAMP even though CRAMP is not responsible for PhoP/PhoQ activation when S. Typhimurium is inside murine macrophages (72).
In sum, CAMPs appear to activate PhoQ by disrupting PhoQ interactions with divalent cations, rather than by PhoQ actually recognizing specific sequences or structural features in CAMPs (76). That a given CAMP activates different PhoP-activated genes to different extents (72) and that some PhoP-activated genes are regulated by other proteins independently of PhoP (77,–80) suggest that certain CAMPs act in a PhoQ-independent fashion.
Mildly Acidic pH
The S. Typhimurium PhoP/PhoQ system is activated by mildly acidic pH (i.e., pH 5) (81). This makes physiological sense given that S. Typhimurium experiences such pH inside macrophage phagosomes (82) and that phoP and phoQ mutants are defective for survival inside macrophages (17, 32). Moreover, phagosome acidification to pH <5.0 is correlated with maximal expression of PhoP-activated genes (83), whereas inhibition of phagosome acidification hinders PhoP/PhoQ activation inside macrophages (83). In addition, acid shock induces specific S. Typhimurium proteins, including PhoP (81), and a mildly acidic pH activates gene transcription in a PhoP-dependent manner (21). Furthermore, the PhoP/PhoQ system is required for the protection of S. Typhimurium from inorganic acid stress and for the production of particular acid-induced proteins (81).
PhoQ had been proposed to sense periplasmic pH because (i) PhoP was phosphorylated more when the periplasmic domain of PhoQ was exposed to a pH of 5.5 than to pH 7.5 in an ex vivo experiment performed with inside-out vesicles reconstituted with the full-length PhoQ (PhoQ’s cytoplasmic domain faced pH 7.5 in both cases); (ii) the purified periplasmic domain of PhoQ exhibited different NMR spectra at different pH values; (iii) the NMR spectrum of a periplasmic PhoQ variant with the H157N substitution at pH 6.5 and pH 3.5 resembled the spectrum of the wild-type PhoQ periplasmic domain experiencing pH 3.5 (84); and (iv) a strain expressing an engineered PhoQ protein with a nonphysiological disulfide bond between W104C and A128C in PhoQ’s periplasmic domain failed to activate PhoP-dependent gene transcription in acidic pH (63). However, it is highly unlikely that PhoQ senses a mildly acidic pH via its periplasmic domain because (i) an S. Typhimurium strain that expresses the full-length PhoQ protein with the H157R substitution instead of the wild-type PhoQ responds to mildly acidic pH like one expressing the wild-type PhoQ protein in vivo (85); (ii) the PhoQ variant with the nonphysiological disulfide bond in the periplasmic domain does actually respond to mildly acidic pH in vivo (85); (iii) the experiment using inside out vesicles lacked critical controls, such as a sensor not regulated by acidic pH (84); (iv) the pH-dependent changes in the NMR spectra of PhoQ’s periplasmic domain are expected for any protein fragment with a large number of protonatable residues; and (v) an S. Typhimurium phoQ mutant that expresses a chimera harboring the periplasmic domain from P. aeruginosa PhoQ and the cytoplasmic domain from S. Typhimurium PhoQ is partially activated by an acidic pH, but it is no longer activated by the antimicrobial peptide C18G (86).
We now know that PhoQ’s response to acidic pH is mediated by PhoQ’s cytoplasmic domain because (i) PhoQ variants with single amino acid substitutions in cytoplasmic residues D233, H409, and Q460 compromised sensing of pH but not of Mg2+ (85); (ii) a PhoQ variant lacking both transmembrane domains and the periplasmic region still responds to mildly acidic pH in vivo (85); (iii) PhoQ is activated in vivo by a decrease in cytoplasmic pH, even if the external pH remains constant (85); and (iv) a chimera consisting of the periplasmic and transmembrane domains of EnvZ and the cytoplasmic domain of PhoQ still responds to mildly acidic pH (85). Taken together, the available data argue that PhoQ senses pH changes via its cytoplasmic domain, which is in contrast to the sensing of Mg2+ and antimicrobial peptides by PhoQ’s periplasmic domain. It is possible that PhoQ does not sense H+ per se but rather a metabolite or physical parameter that changes when the cytoplasmic pH decreases.
The response to mildly acidic pH by PhoQ appears to be an evolved state in both pathogenic S. Typhimurium and commensal E. coli because full PhoQ activation by mildly acidic pH requires additional, distinct, horizontally acquired proteins in both species. In S. Typhimurium, the PhoP-activated horizontally acquired ugtL gene specifies an inner membrane protein that binds to PhoQ, stimulating PhoQ’s ability to autophosphorylate from ATP in vitro and to generate PhoP-P in vivo (87) (Fig. 3A). UgtL is required for PhoQ’s response to a mildly acidic pH but dispensable when the activating signal is low Mg2+ (87). UgtL furthers PhoP phosphorylation even when expressed under noninducing conditions (87), indicating that UgtL can operate independently of mildly acidic pH sensing by PhoQ. In addition to ugtL, mildly acidic pH activation of the S. Typhimurium PhoP/PhoQ system requires the horizontally acquired ssrB gene, which specifies a protein that binds to the ugtL promoter, thus overcoming the silencing effect exerted by the H-NS protein, and directly promotes transcription of the phoP gene (88) (Fig. 3A).
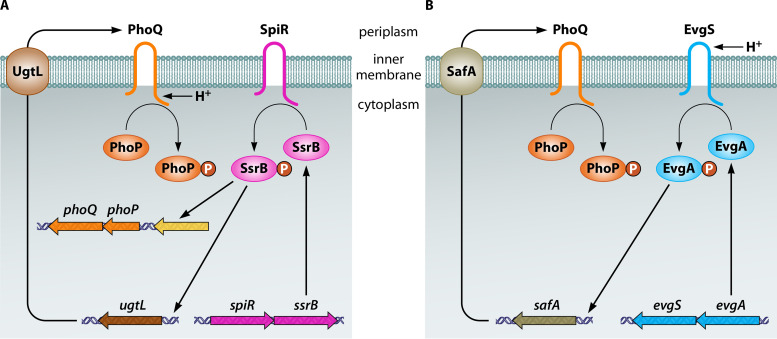
Activation of the PhoP/PhoQ system by mildly acidic pH in S. Typhimurium versus E. coli. (A) In S. Typhimurium, PhoQ utilizes its cytoplasmic domain to detect mildly acidic pH (or something that changes upon a decrease in the cytoplasmic pH). Full activation requires binding of the UgtL protein to PhoQ, which stimulates PhoQ autophosphorylation. It also requires the regulator SsrB to promote transcription of both the ugtL gene and the phoPQ operon; the latter results from SsrB binding to the upstream coding region (in yellow). (B) In E. coli, the sensor EvgS responds to mildly acidic pH by activating its cognate regulator EvgA, which, in turn, promotes transcription of the safA gene. The SafA protein binds to PhoQ and promotes PhoQ autophosphorylation. Not shown is the positive feedback that PhoP exerts on the phoPQ operon in both species and on the ugtL gene in S. Typhimurium.
In E. coli, PhoQ activation by mildly acidic pH requires the periplasmic protein SafA, which promotes PhoQ phosphorylation (89) (Fig. 3B). Like UgtL in S. Typhimurium, SafA is dispensable for PhoQ activation by low Mg2+ in E. coli (90). This is further supported by the fact that the SafA-mediated PhoQ activation relies on a periplasmic sensor pocket that is distinct from the region involved in sensing Mg2+ (91). In addition, safA was also acquired by horizontal gene transfer. However, unlike ugtL, safA is not regulated by PhoP or SsrB but rather by EvgA (89) (Fig. 3B), a regulator that forms a two-component system with the sensor EvgS (92). Encoded by horizontally acquired genes, the EvgS/EvgA system responds to a decrease in pH by promoting SafA expression, thus allowing PhoQ activation by acidic pH (90, 93). That E. coli phoP mutants are hypersensitive to killing by acidic pH (11) provides a physiological explanation for acidic pH being a signal activating the E. coli PhoP/PhoQ system.
Acidic pH sensing is largely independent of divalent cation sensing because the effects of low Mg2+ and low pH are cumulative (84) and also because several PhoQ cytoplasmic residues (i.e., D233, H409, and Q460) have been linked specifically to pH sensing (85). That PhoQ senses cytoplasmic pH and that the response to mildly acidic pH requires the PhoP-activated UgtL protein imply a delay between a pH change in S. Typhimurium’s surroundings and PhoP activation in the cytoplasm. This delay may reflect a need to respond to prolonged extracellular acidification for changes in cytoplasmic pH to take place. Moreover, it is in contrast to pH sensing by sensors such as PmrB, which takes place in the periplasm and does not require genes regulated by PmrB’s cognate regulator PmrA (94). The kinetics of PhoP/PhoQ activation by acidic pH are expected to be slower than those triggered by low Mg2+ in both S. Typhimurium and E. coli, but, as discussed above, for different reasons (Fig. 3).
S. bongori, the largely nonpathogenic species of the genus Salmonella (95), activates the PhoP/PhoQ system in response to low Mg2+ and C18G, like the pathogenic serovar S. Typhimurium. However, it is defective for activation in mildly acidic pH (85) because the cytoplasmic PhoQ residues implicated in the response to acidic pH in S. Typhimurium are not conserved in the S. bongori protein (85) and also because S. bongori lacks both the ssrB gene and the SsrB binding site in the ugtL promoter, which prevents UgtL production under the conditions that normally activate the SsrB protein (88). Curiously, the S. bongori ugtL gene does complement a S. Typhimurium ugtL mutant when expressed from a heterologous promoter (88). This is in spite of the fact that the UgtL proteins of S. bongori and S. Typhimurium share only 55% amino acid identity, much lower than most proteins present in the two organisms (e.g., their PhoQ proteins share 99% amino acid identity). These results suggest that UgtL operates in S. bongori but under different conditions than in S. Typhimurium.
Hyperosmotic Stress
An osmotic upshift resulting from treatment with 300 mM NaCl causes a transient upregulation of the PhoP/PhoQ system in E. coli carrying its own phoQ gene or the one from S. Typhimurium (96). This upregulation is fast, taking place within minutes, but transient, with a return to prestimulating conditions within 15 min. PhoP activation by osmotic upshift is independent of OmpR/EnvZ, a two-component system that responds to changes in osmolarity (97). The osmoregulation of PhoP/PhoQ makes physiological sense given that phoP and phoQ mutants exhibit a longer lag phase than the wild-type strain when exposed to osmotic stress (96). The PhoP-mediated resistance to osmotic stress is mediated, in part, by the PhoP-activated iraM gene (96), which encodes a protein that binds to the protease adaptor RssB, thereby preventing RssB from delivering the alternative sigma factor RpoS (also referred to as sigma S) to the protease ClpXP for degradation (98) (discussed below under “PhoP Controls Protease Activity and Specificity”). (RssB is also referred to as SprE in E. coli and MviA in S. Typhimurium.)
The sensor PhoQ responds to both ionic and nonionic osmolytes, such as sucrose and sorbitol (96). Osmosensing takes place even in the presence of repressing (i.e., 10 mM) concentrations of Mg2+ and in the absence of PhoQ’s periplasmic domain, which, as discussed above, is essential for sensing divalent cations and CAMPs (96). PhoQ uses its transmembrane domains to detect an osmotically perturbed membrane (96), in contrast to pH sensing in the cytoplasm (85, 87) and Mg2+ sensing in the periplasm (18). Curiously, the mRNA leader regions of the PhoP-activated mgtA gene (99) and mgtCBRU-cigR operon (100) from S. Typhimurium respond to hyperosmotic stress by favoring transcription elongation into their respective coding regions.
Periplasmic Redox State
In E. coli and S. Typhimurium, the periplasmic protein DsbA is responsible for disulfide bond formation (101). Genetically speaking, DsbA represses expression of PhoP-activated genes in a manner dependent on the sensor PhoQ, its negative regulator MgrB, and the transcriptional activator PhoP (102, 103). The PhoP-activated gene mgrB (104) encodes a small integral membrane protein that stimulates PhoP-P dephosphorylation by PhoQ (105). DsbA is proposed to favor MgrB activity by stimulating disulfide bond formation among the three cysteines in MgrB, two of which are located in the periplasm (103). This is because the PhoP/PhoQ system is derepressed upon inactivation of the dsbB gene, in the presence of the reducing agent dithiothreitol, or upon substitution of the periplasmic cysteines by alanines (103). (The dsbB gene encodes a protein required for oxidizing reduced DsbA [101].) These findings argue that the MgrB protein provides the means to incorporate information about the oxidative status of the bacterium’s periplasm into the activation status of the PhoQ protein (102).
Long-Chain Unsaturated Fatty Acids
Several long-chain unsaturated fatty acids, including linoleic, linolenic, and palmitoleic acids, downregulate transcription of PhoP-activated genes 2- to 4-fold when added to S. Typhimurium growing in Luria-Bertani (LB) broth. This downregulation is less than that achieved by >2 mM Mg2+ (106). The autokinase activity of the PhoQ protein present in vesicles decreased if linoleic acid was added during bacterial growth, but curiously, not if added to PhoQ-enriched vesicles prepared from bacteria grown in media lacking linoleic acid (106). The autokinase activity of the sensor EnvZ was impervious to the presence of linoleic acid, suggesting that this fatty acid does not target the regions of sequence similarity between PhoQ and EnvZ (106). Despite containing linoleic acid (107), bile does not regulate transcription of PhoP-activated promoters (108). The periplasmic domain of PhoQ is sufficient to achieve binding to linoleic acid in vitro (109), suggesting that this domain senses fatty acids. However, as discussed above for PhoQ activation by CAMPs, the large size of these molecules compared to Mg2+ suggests a distinct mechanism of interaction with PhoQ. This notion is further supported by the different NMR spectra displayed by the PhoQ periplasmic domain when bound to Mg2+ versus linoleic acid (109). The in vivo binding of short chain unsaturated fatty acids and the relevance of such binding remain to be determined. This is because a variety of factors, not all related to ligand binding, can affect NMR spectra and also because the in vivo behavior of strains expressing PhoQ variants with substitutions in amino acids perturbed by short-chain unsaturated fatty acids is yet to be reported.
Acetate
Acetate decreases PhoQ’s activation of PhoP in both S. Typhimurium and E. coli growing in defined media in the absence of added Mg2+ (110). Following its uptake into the cytoplasm, acetate is converted into acetyl coenzyme A (acetyl-CoA), which exerts its inhibitory effects even in a strain lacking PhoQ’s periplasmic domain. Therefore, changes in the extracytoplasmic abundance of a molecule are detected by the effects that a metabolite of that molecule exerts on the cytoplasmic domain of PhoQ. Acetyl-CoA acts as a noncompetitive inhibitor of PhoQ autophosphorylation in vitro, but only at concentrations twice as high as those achieved in vivo upon growth in acetate concentrations that inhibit the PhoP/PhoQ system (110).
Acetate availability does alter the amount of active PhoP protein. However, acetate appears to operate independently of PhoQ. First, the increase in acetyl CoA taking place during growth in acetate may directly inactivate PhoP, which is acetylated at three positions (111,–113) (see below under “PhoP Acetylation”). And second, an increase in the cytoplasmic concentration of acetyl-CoA decreases PhoP proteolysis by promoting acetylation of HspQ (114), an anti-adaptor for ClpS, the protease adaptor that targets PhoP for degradation by the protease ClpAP (58).
In sum, PhoQ responds to a variety of activating and repressing chemical signals as well as to changes in osmolarity by modifying the amount of PhoP-P. These signals are detected by different PhoQ domains and sometimes require additional proteins to exert their regulatory effects.
SIGNALS ACTING ON PhoP: IMPACT OF METABOLISM AND CELL ENVELOPE STRESS ON AMOUNTS OF ACTIVE PhoP PROTEIN
The majority of the signals impacting PhoP-dependent gene transcription are processed by PhoQ, which is responsible for altering PhoP’s phosphorylated status. However, certain signals do act on PhoP and impact expression of PhoP-regulated genes.
PhoP Acetylation
The S. Typhimurium PhoP can be acetylated at three lysine residues. Acetylation of K201 is carried out by the protein acetyl transferase Pat using acetyl-CoA as an acetyl donor (112). Because K201 is located in PhoP’s DNA binding region, acetylation of K201 impairs PhoP binding to DNA, resulting in a 2- to 5-fold decrease in transcription of PhoP-activated genes (112). However, acetylation of K201 does not interfere with in vitro PhoP autophosphorylation from acetyl phosphate at D52 (112), the conserved aspartate site of phosphorylation in regulators of the two-component system family (50). K201 is highly conserved not only in other PhoP sequelogs (115) but also in regulators from the two-component system family encoded in the Staphylococcus aureus and Pseudomonas aeruginosa genomes (112). This conservation raises the possibility of other regulators, like the S. Typhimurium PhoP protein, being acetylated by Pat.
Metabolic signals may control PhoP acetylation because acetyl-CoA abundance changes with the carbon source in which a bacterium is grown. For example, there is more acetyl-CoA when organisms are grown in glucose than in glycerol as primary carbon source (116). In addition, deacetylation of PhoP at K201 is carried out by the deacetylase CobB in an NAD+-dependent manner (112), suggesting that conditions altering NAD+ abundance contribute to PhoP’s acetylation status.
Environments that promote PhoP phosphorylation, such as growth in low Mg2+ or at a mildly acidic pH, decrease PhoP acetylation at K201 (112), raising questions about the physiological role that PhoP acetylation at K201 plays. That is to say, PhoP acetylation and phosphorylation are favored by opposite signals. Therefore, one possibility is for acetylated PhoP to have a distinct biochemical activity (and thus physiological function) from that exhibited by acetylated PhoP-P, one that would not entail DNA binding, at least not via PhoP’s canonical DNA-binding domain. The fraction of the total PhoP protein that is acetylated at K201 increased 2-fold in bacteria grown in LB medium in the absence of added Mg2+ versus LB medium with a repressing (10 mM) concentration of Mg2+ (112), and it decreased 2-fold when growth was compared at pH 7.0 versus pH 5.0 (112), suggesting that conditions that favor PhoP phosphorylation at D52 (85, 87) reduce PhoP acetylation at K201.
In addition to Pat-dependent acetylation of K201, PhoP self-acetylates (i.e., nonenzymatically) from acetyl phosphate at K102 (111) and K88 (113). Acetylation at K102 increases upon medium supplementation with glucose or acetate, carbon sources that elevate the concentration of acetyl phosphate, and it decreases some 2-fold under conditions that favor PhoP phosphorylation, such as growth in low Mg2+ or at pH 5.0 (relative to growth in mM Mg2+ and pH 7.7, respectively) (111). This is in spite of the fact that growth in low Mg2+ favors protein acetylation from acetyl phosphate in E. coli (117). As discussed above for acetylation of K201, K102 acetylation is observed in ~50% of the PhoP molecules (111). Acetylation of K102 decreases PhoP-dependent gene transcription even though K102 is located within PhoP’s receiver domain (118). This effect may result from the inhibition of PhoP phosphorylation at D52 when K102 is acetylated (111) and from phosphorylation at D52 favoring PhoP dimerization, which is the form of PhoP that binds DNA and regulates gene transcription (14). PhoP variants with amino acid substitutions in K102 mimicking the acetylated and nonacetylated states were equally defective in gene transcription (111), indicating that such changes at K102 compromise PhoP activity as a transcriptional regulator.
Substitution of the PhoP lysines at positions 201 and 102 by arginines results in different consequences even though the K-to-R substitution is expected to mimic the nonacetylated state. That is, the PhoP K201R protein retained a wild-type ability to promote transcription of PhoP-activated genes, whereas the PhoP K102R variant was not functional (111).
The PhoP fraction acetylated at K88 decreases 3-fold by 24 h after S. Typhimurium internalization by macrophages compared to before infection (113). Acetylation at K88 decreases PhoP dimerization, DNA binding, and PhoP’s ability to activate gene transcription (113). It is presently unclear what fraction of the total PhoP protein is acetylated solely at K88, K102, and K201 or in various combinations of double- or triple-phosphorylated forms at these lysines.
The Sigma E-Activated sRNA MicA Represses Expression of the PhoP Protein
The sRNA MicA is predicted to base pair with the translation initiation region of the phoP mRNA in several enteric bacteria, including E. coli, Salmonella, and Enterobacter, thereby decreasing phoP translation (119). When MicA is overexpressed, transcription of PhoP-activated genes decreases ~3-fold. The alternative sigma factor sigma E is liberated from its inactive inner-membrane form when bacteria experience specific outer-membrane stresses (120). Because micA is a sigma E-activated gene, stress conditions that liberate sigma E would likely decrease PhoP amounts. However, expression of PhoP-activated genes in a micA mutant was not significantly different from that displayed by wild-type E. coli under normal inducing conditions for sigma E (119), arguing that the effects of MicA are modest, at least under the reported conditions.
FEEDBACK LOOPS CONTROLLING PhoP-P PROTEIN AMOUNTS
S. Typhimurium employs several regulatory mechanisms to modify PhoP-P amounts, which change continuously even under constant inducing conditions. The total number of PhoP monomers present in S. Typhimurium has been estimated at 1,600 under noninducing conditions and 10,000 following 60 min in low (10 μM) Mg2+ defined medium conditions (3). In the case of PhoQ, the number of PhoQ monomers per cell increases from ~300 under noninducing conditions to ~650 under inducing conditions (3). Thus, the ratio of PhoP-to-PhoQ molecules increases from ~7 to >18 between noninducing and 60 min after inducing conditions. The latter ratio is about half of the OmpR-to-EnvZ (121) and PhoP-to-PhoQ (122) ratios in E. coli during steady-state growth in complex media, which activates the PhoP/PhoQ system but not as much as growth in defined medium with 10 μM Mg2+ (123).
The regulatory mechanisms discussed below are mediated by PhoP-regulated gene products, operating only when the PhoP/PhoQ system is active. Thus, they differ from other mechanisms controlling the abundance, phosphorylation, and acetylation carried out by proteins and metabolites not known to be regulated by PhoP.
Transcriptional Autoregulation of the PhoP/PhoQ System
The S. Typhimurium phoPQ operon is transcribed from two promoters (124). The first is a constitutive promoter that provides the basal amount of PhoQ protein necessary to detect changes in conditions and of PhoP protein required to initiate a response to such changes. The second promoter is directly activated by PhoP-P (Fig. 4). Expression from the latter promoter is necessary for S. Typhimurium to express PhoP-activated genes at the levels necessary for a normal physiological response to stress, including the successful infection of a mammalian host (15). Positive feedback of PhoP on the phoPQ operon is necessary for S. Typhimurium to grow in low Mg2+ because a mutant lacking the PhoP binding site (designated PhoP box) in the promoter of the phoPQ operon was as defective for growth in low Mg2+ media as a mutant lacking the phoP and phoQ genes (28). The E. coli phoPQ operon is also transcribed from two promoters. However, the phoPQ operons of the two species differ in that the positively autoregulated promoter is distal to the coding region in S. Typhimurium but close to it in E. coli (104, 124).
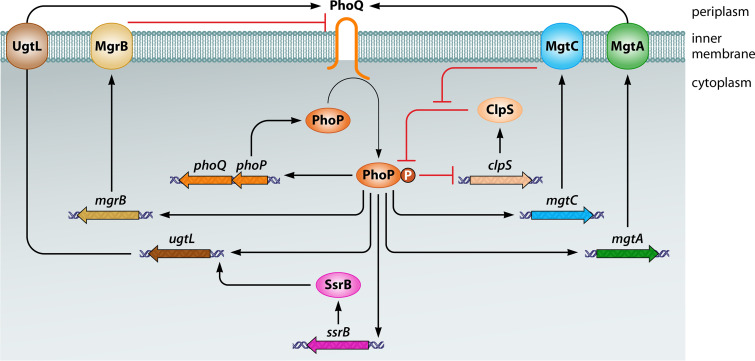
Feedback mechanism controlling the abundance of active PhoP protein in S. Typhimurium. PhoP-P is a direct transcriptional activator of the phoPQ operon and mgtA, mgtC, ugtL, mgrB, and ssrB genes, and a direct transcriptional repressor of the clpS gene. MgtA increases PhoQ-dependent phosphorylation of PhoP by decreasing Mg2+ amounts in the periplasm. MgtC increases the amounts of PhoP and PhoP-P by preventing proteolysis of both forms of PhoP by the ClpAP protease in a ClpS-dependent manner. The UgtL and MgrB proteins stimulate and inhibit PhoQ autophosphorylation, respectively. SsrB promotes ugtL gene transcription. Some of these mechanisms also operate in bacterial species other than S. Typhimurium. Not shown is that SsrB binds to the purB coding region upstream of phoPQ, promoting transcription of this operon, or that PhoP promotes transcription of the qad gene, which specifies an acetyl-CoA binding protein necessary for acetylation of HspQ, an anti-adaptor of the protease adaptor ClpS.
In Pseudomonas aeruginosa, the phoP and phoQ genes are preceded by the oprH gene (125), which forms a three-gene operon positively regulated by PhoP but negatively regulated by PhoQ (125). That the oprH gene is part of the P. aeruginosa PhoP regulon makes physiological sense given that it specifies an outer membrane porin which upon overexpression confers bacterial survival toward EDTA (126), a chelator of Mg2+ and other divalent cations. The DNA region separating the oprH and phoP coding regions is 79 nucleotides (nt) long, raising the possibility of this region harboring a promoter that provides the basal levels of transcription of the phoP and phoQ genes and/or being targeted by a regulatory protein or sRNA.
In Yersinia pestis, the phoP and phoQ genes are also part of a three-gene operon positively regulated by PhoP (28). Like the oprH gene in P. aeruginosa, the y1795 gene precedes phoP in Y. pestis. Although both OprH and Y1795 are outer membrane proteins, they do not share amino acid sequence identity. The organization of the latter two phoPQ-containing operons is reminiscent of the S. Typhimurium pmrCAB operon, in which the pmrA and pmrB genes specify a two-component system (127). A constitutive promoter located within the pmrC coding region of the PmrA-activated pmrCAB operon provides basal transcription of the pmrA and pmrB genes (40).
Transcriptional autoregulation is a property that the PhoP/PhoQ system shares with several two-component systems (128) and, more generally, with other DNA-binding transcriptional activators (129). However, the positive feedback that PhoP-P exerts on its own promoter does not appear to reduce the response to an inducing signal, as has been reported for regulators that belong to protein families other than the two-component system family (129). Likewise, it does not result in bistability in a wild-type strain (130). Bistability in the PhoP/PhoQ system has been observed in a strain harboring a single point mutation that abolishes PhoQ’s phosphatase activity towards PhoP-P (131).
Intrinsic PhoQ Feedback Alters PhoP-P Amounts under Constant Inducing Conditions
When Salmonella is switched from repressing to activating Mg2+ concentrations for the PhoP/PhoQ system (e.g., from 10 mM to <50 μM Mg2+), the amount of PhoP-P increases, reaches a peak at 20 to 30 min postinduction, and then decreases to about one-third of the maximum (15), which is puzzling given the continuous presence of inducing conditions. This surge in PhoP-P amounts is accompanied by increases in promoter occupancy by PhoP-P and in mRNA abundance of PhoP-activated genes (15). The surge requires PhoP-P to positively activate its own transcription because an engineered strain that transcribed the phoP gene constitutively achieved steady-state levels of PhoP-activated mRNAs monotonically, that is, without the surge (15). The surge is critical for S. Typhimurium virulence because the engineered strain was as attenuated for virulence in mice as a strain lacking the phoP or phoQ genes (15).
What, then, promotes a decrease in PhoP-P amounts when S. Typhimurium is continuously experiencing inducing conditions for the sensor PhoQ? The decrease in PhoP-P from peak values requires PhoQ’s phosphatase activity toward PhoP-P (15). The surge was recapitulated in vitro using purified PhoP and PhoQ proteins, ATP, and adenosine diphosphate (ADP) (3), arguing against an additional factor (i.e., a PhoP-activated gene product) being necessary for the surge. ADP stimulation of PhoQ’s phosphatase activity is critical for dephosphorylation of PhoP-P because PhoQ variants defective in holding on to ADP exhibited sustained PhoP-P amounts, unlike reactions carried out with the wild-type PhoQ protein (3).
The regulator PmrA exhibits an analogous surge in PmrA-P when S. Typhimurium experiences inducing signals for the sensor PmrB in vivo. Moreover, like the PhoP/PhoQ system, the PmrA-P surge was reconstituted in vitro using purified PmrA and PmrB proteins, ATP, and ADP (3). These results strongly suggest that a common mechanism is responsible for the surge across two-component systems in which ADP stimulates the phosphatase activity of the sensor towards its cognate phosphorylated regulator. Because ATP and ADP bind to the same nucleotide-binding pocket in sensor proteins, ADP binding not only stimulates dephosphorylation of the phosphorylated regulator but also prevents ATP binding, thereby decreasing sensor autophosphorylation (3). This is in contrast to an alternative model (discussed below under “The PhoP-Activated MgrB and SlyB Proteins Exert Negative Feedback on PhoP/PhoQ”) in which each two-component system controls expression of a protein(s) that exclusively exerts negative feedback on the system that controls its expression. Mathematical models of the PhoP-P surge provide an understanding of the various parameters under which the surge takes place (3, 132).
The PhoP-Activated MgrB and SlyB Proteins Exert Negative Feedback on PhoP/PhoQ
The PhoP-activated mgrB gene specifies a 47 amino acid inner membrane protein that exerts negative feedback on PhoP by binding to PhoQ (105) (Fig. 4). In E. coli, mgrB inactivation increases transcription of PhoP-activated genes, whereas mgrB overexpression decreases it (105). These in vivo effects were observed during growth at both inducing (100 μM) and repressing (10 mM) Mg2+ concentrations (105), raising the possibility of mgrB also being transcribed from a PhoP-independent promoter given that 10 mM Mg2+ is a repressing condition for PhoQ. MgrB also exerts its repressing effects when the PhoQ-inducing conditions are mildly acidic pH or C18G, indicating that MgrB acts differently from other PhoP-regulated gene products that operate in a PhoQ-inducing, condition-dependent manner (57) (see below under “The Mg2+ Transporter MgtA Increases PhoP-P amounts in a PhoQ-Dependent Manner,” “The MgtC Protein Protects PhoP and PhoP-P from Proteolysis by ClpSAP in Multiple Ways,” “PhoP Reduces the Amounts of Active Protease Adaptor ClpS,” “EIIANtr Inhibits PhoP Binding to DNA and Is Degraded by Lon in a PhoP-Dependent Manner,” and “PhoP Promotes Transcription of DNA-Binding Protein SsrB and Vice Versa”). MgrB’s inhibitory activity requires PhoQ residues located in the cytoplasm, transmembrane domain, and periplasm (133). Although feedback mechanisms often control expression kinetics and cell-to-cell expression behavior (134), wild-type and mgrB strains behaved alike in these two properties (105). The in vivo behavior of PhoP-activated genes in wild-type versus mgrB mutant suggests that the MgrB protein inhibits PhoQ’s autophosphorylation but may also stimulate PhoQ’s phosphatase activity toward PhoP-P (135).
Mutations in the mgrB gene have been repeatedly recovered in Klebsiella pneumoniae isolates exhibiting resistance to the antibiotic colistin (136,–138). This is true for mutants isolated in the laboratory as well as in clinical strains of this pathogen. The mgrB mutants display upregulation of the PhoP/PhoQ system and of its regulated targets, the most relevant being the pmrFIJKLM operon because of its role in the chemical modification of the LPS, resulting in colistin resistance (36, 139). Likewise, an insertion sequence decreasing mgrB expression in a Klebsiella oxytoca clinical isolate conferred colistin resistance (140). By contrast, there are no reported mgrB mutations conferring colistin resistance in Enterobacter and Citrobacter (141), suggesting that regulation of phoP by mgrB and/or regulation of colistin resistance by PhoP is not conserved in either of the latter two species.
The decrease in PhoP-P taking place following the surge in PhoP-P (15) has been ascribed to the inhibitory activity that MgrB exerts on PhoQ (135) rather than to an intrinsic property of the PhoQ protein, as discussed above (3). The different conclusions reached by two groups may reflect the use of different bacterial species (E. coli versus S. Typhimurium), growth conditions (agarose pads at 30°C versus shaking liquid media at 37°C), measured output (indirect [fluorescence from plasmid-based promoter fusions to reporter genes] versus direct [mRNA abundance of PhoP-activated genes and occupancy by the PhoP-P protein of the chromosomal copies of PhoP-activated promoters]), or Mg2+ concentration of the starting culture (50 mM versus 10 mM) in the in vivo experiments. These significant differences may account for the minimal adaptation observed in the experiments carried out with E. coli compared to S. Typhimurium. In addition, the two groups used different PhoQ constructs for the in vitro experiments: His6-MBP-tagged E. coli PhoQ lacking both transmembrane domains and the periplasmic region versus Strep-tagged full-length S. Typhimurium PhoQ protein. The former PhoQ was proposed to carry out multiple rounds of autophosphorylation from ATP in the absence of PhoP, leading to the notion that ADP does not remain in the nucleotide binding pocket (135). By contrast, the latter PhoQ was not dephosphorylated after incubation with ATP (3), resulting in the proposal that ADP remains in the nucleotide binding pocket, where it stimulates PhoQ’s phosphatase activity towards PhoP-P.
In S. Typhimurium, the MgrB protein inhibits PhoQ protein autophosphorylation independently of the PhoQ autophosphorylation stimulator UgtL when the inducing signal is mildly acidic pH (87). Likewise, UgtL stimulates PhoQ autophosphorylation even in an mgrB mutant (87). This provides a singular example of two proteins encoded by genes directly activated by PhoP exerting opposite activities on PhoP’s cognate sensor PhoQ.
The PhoP-activated slyB gene encodes a putative lipoprotein that exerts mild negative feedback on the PhoP/PhoQ system because PhoP-activated genes are derepressed slightly in a slyB-null mutant and also because slyB overexpression from a heterologous promoter inhibits S. Typhimurium growth in low-Mg2+ media (28). Like mgrB, slyB is also transcribed under nonactivating conditions for PhoQ, thereby establishing a threshold that must be surpassed for PhoQ to activate PhoP.
The Mg2+ Transporter MgtA Increases PhoP-P Amounts in a PhoQ-Dependent Manner
The PhoP-activated mgtA gene specifies a Mg2+ transporter that imports Mg2+ from the periplasm into the cytoplasm (35, 142, 143). The MgtA protein increases PhoP-P amounts by removing Mg2+ from the periplasmic space, where it inhibits PhoQ activity (57) (Fig. 4). The increase in PhoP-P enables transcription of those promoters that require large amounts of PhoP-P for transcription to take place (57).
Expression of a subset of the genes directly regulated by PhoP is delayed until the MgtA protein is made (57). This is because the mgtA mRNA includes an unusually long leader region that functions as a Mg2+-responding riboswitch and determines whether RNA polymerase continues transcription into the mgtA coding region or stops within the leader (144, 145). When the Mg2+ concentration drops below a certain threshold, the mgtA coding region is transcribed, the corresponding mRNA is translated, and the resulting MgtA protein imports Mg2+, which is believed to activate the PhoQ protein by reducing the abundance of periplasmic Mg2+ (57). (A discussion of the proposed mechanisms of Mg2+ sensing by the mgtA leader is available [146].)
MgtA exerts positive feedback on PhoQ by virtue of being a Mg2+ importer (57), as opposed to directly binding to the PhoQ protein as UgtL does (87) (Fig. 4) or as other transporters that bind to membrane-integrated sensors do (147). Moreover, MgtA requires PhoQ’s ability to sense periplasmic Mg2+ because mgtA inactivation had no effect on expression of PhoP-activated genes in a strain with a PhoQ protein blind to silencing by Mg2+ (57). Likewise, PhoP-dependent gene transcription was unaltered upon mgtA inactivation when the PhoQ-inducing signal was C18G or a mildly acidic pH (57), since MgtA is not known to transport C18G or to alter cytoplasmic pH. In sum, MgtA appears to activate PhoQ by removing Mg2+ from the periplasm, thus allowing PhoQ to promote PhoP phosphorylation; however, MgtA-dependent changes in the concentration of free periplasmic Mg2+ have not actually been reported.
When the PhoQ-inducing signal is low Mg2+, the positive feedback exerted by MgtA allows S. Typhimurium to differentially express PhoP-regulated genes as a function of time: MgtA-independent genes are expressed before those that are MgtA dependent. In addition, the MgtA feedback enables different PhoQ-activating conditions (i.e., low periplasmic Mg2+ versus mildly acidic pH) to elicit different gene expression outputs at a given time (57). The PhoP-activated genes most strongly regulated in an mgtA-dependent manner are the horizontally acquired pagC, pagD, pagK, pgtE, and pcgL genes (57), which encode proteins with a variety of biochemical activities.
MgtA exerts positive feedback by removing an inhibitory signal from the signal transduction system it activates. Moreover, it implements an expression hierarchy within the PhoP regulon in a manner different from that mediated by a classical transcriptional cascade, in which one regulator controls expression of another regulator that is then responsible for expression of a structural gene(s) (148).
The MgtC Protein Protects PhoP and PhoP-P from Proteolysis by ClpSAP in Multiple Ways
The PhoP protein is a ClpS-dependent substrate of the ClpAP protease (58) (Fig. 4). That is to say, the protease adaptor ClpS recognizes PhoP and delivers it to the chaperone ClpA for degradation in the proteolytic chamber of ClpP. Like other members of the ATPases associated with multiple activities (AAA+) protein family (149), ClpA utilizes the energy derived from ATP hydrolysis to unfold its substrates. Therefore, conditions that alter the amounts of ATP (37) or the amounts or availability of ClpS (16, 58, 114) impact PhoP stability.
When experiencing low cytoplasmic Mg2+, S. Typhimurium utilizes multiple mechanisms to hinder PhoP degradation by the protease ClpSAP. Two of these mechanisms are directly mediated by the PhoP-activated mgtC gene, which specifies an integral membrane protein that binds the PhoP protein and inhibits ATP synthesis (150) by reducing phosphate uptake (151) and by binding to the F1F0 ATPase (150). MgtC reduces the ATP concentration below the threshold necessary for normal proteolysis of PhoP (58) and other functional proteins targeted by ATP-dependent proteases (37). MgtC binding to PhoP prevents ClpS access to PhoP (Fig. 4), thereby sparing PhoP proteolysis by ClpSAP (58). MgtC’s activity as protector of a protease substrate differs from that of anti-adaptors that sequester protease adaptors, thereby sparing multiple substrates of a given adaptor (152). Because MgtC acts stoichiometrically, PhoP stability is determined by the relative abundance of ClpS, F1F0 ATPase, PhoP, and MgtC (58), the affinities of MgtC and ClpS for their distinct interacting partners, and the amounts of CigR and MgtR. The CigR protein sequesters MgtC, thereby preventing MgtC from reducing ATP amounts and protecting PhoP (153). MgtR is an adaptor of the protease FtsH necessary for MgtC proteolysis (154).
When S. Typhimurium first experiences a low Mg2+ environment, PhoP promotes transcription from the mgtC promoter, but the MgtC protein is not produced. This is because the unusually long mgtC leader prevents transcription elongation into the mgtC coding region until specific cytoplasmic conditions are met (100, 144, 155, 156). Under these conditions, PhoP protein amounts increase (15) because PhoP promotes its own transcription (124). However, some PhoP protein is bound by ClpS and degraded by ClpAP (58). Once transcription elongation into the mgtC coding region takes place, the synthesized MgtC protein must surpass CigR protein amounts (153) for a reduction in ATP synthesis and protection of PhoP and PhoP-P from ClpSAP to take place (58). Because MgtC has a higher affinity for PhoP than does ClpS, once MgtC amounts reach a certain threshold, MgtC sequesters PhoP away from ClpS, resulting in PhoP accumulation (58) and PhoP-P binding to target DNAs.
The positive feedback that MgtC exerts on its transcriptional activator PhoP (i.e., a 4-fold increase in PhoP’s half-life) (Fig. 4) allows S. Typhimurium to delay transcription of a subset of PhoP-activated genes until the cytoplasmic conditions resulting in MgtC production are met (58). As discussed above for the MgtA protein (57), the PhoP-activated genes whose expression is most affected by MgtC-dependent stabilization are horizontally acquired (58).
PhoP Reduces the Amounts of Active Protease Adaptor ClpS
PhoP hinders its own proteolysis by two additional mechanisms. First, PhoP binds to the clpS promoter and represses clpS transcription directly (16) (Fig. 4), which reduces ClpS protein amounts. This repression is mgtA and mgtC dependent (16), indicating that PhoP binding to the clpS promoter requires large PhoP-P amounts achieved only after MgtC accumulates to levels that protect PhoP from proteolysis by ClpSAP and MgtA imports Mg2+ from the periplasm, thereby favoring PhoQ phosphorylation of PhoP.
Second, PhoP is a direct transcriptional activator of qad (157), a gene specifying an acetyl-CoA binding protein necessary for acetylation of HspQ (114), an anti-adaptor for ClpS (114). Acetylation protects HspQ from degradation by the Lon protease (114). The accumulated HspQ binds to ClpS and inhibits ClpS-dependent proteolysis (114). Therefore, conditions favoring acetyl-CoA accumulation enhance HspQ acetylation (114), resulting in decreased PhoP proteolysis by ClpSAP (114, 157). Like the PhoP repression of clpS transcription, PhoP activation of qad transcription is mgtA and mgtC dependent (157).
In sum, PhoP utilizes two independent strategies to reduce the amount of ClpS protein available to bring PhoP to ClpAP for degradation. On the one hand, PhoP repression of clpS transcription reduces the synthesis of new ClpS protein. On the other hand, PhoP activation of qad transcription reduces the availability of preexisting ClpS protein by stabilizing ClpS anti-adaptor HspQ.
When S. Typhimurium experiences low cytoplasmic Mg2+, PhoP repression of clpS transcription is necessary for full transcription of some PhoP-activated genes (16). An engineered strain with a mutant clpS promoter no longer repressible by PhoP harbors smaller amounts of PhoP protein and of PhoP-activated mRNAs than the isogenic wild-type strain (16). The PhoP protection from ClpSAP exerted by MgtC has a stronger effect on the abundance of both PhoP protein and PhoP-activated mRNAs than PhoP repression of clpS transcription (16). E. coli, which lacks mgtC (158), retains PhoP’s ability to repress clpS transcription (16) and to promote qad transcription (157) (Fig. 4). Therefore, closely related bacterial species differ in some of the feedback mechanisms controlling the abundance of active PhoP protein, which may contribute to the species-specific behaviors mediated by conserved genes.
EIIANtr Inhibits PhoP Binding to DNA and Is Degraded by Lon in a PhoP-Dependent Manner
EIIANtr is a component of the nitrogen-metabolic phosphotransferase system that inhibits PhoP’s ability to bind DNA and to promote transcription of PhoP-activated genes in S. Typhimurium (159). PhoP, in turn, promotes EIIANtr proteolysis by Lon via an unknown mechanism (159), establishing a double-negative feedback loop that controls PhoP amounts in response to the signals that govern EIIANtr abundance. EIIANtr controls PhoP independently of EIIANtr’s phosphorylation status. The inhibitory effect on PhoP is specific to EIIANtr because its paralog EIIAGlc has no effect on PhoP binding to DNA (159). Moreover, EIIANtr appears to act specifically on PhoP because it did not hinder PmrA binding to its target DNA. EIIANtr exerts its effects under different PhoQ-inducing conditions. The increase in EIIANtr abundance taking place under noninducing conditions for PhoP/PhoQ helps organisms rapidly turn off PhoP/PhoQ.
EIIANtr delays transcription of PhoP-activated genes when S. Typhimurium is inside macrophages, thereby establishing a threshold that the PhoP protein must cross to exert its regulatory effects (159). The EIIANtr effect on PhoP is reminiscent of the threshold the CigR protein establishes for the MgtC protein (153). However, these proteins differ not only in their targets but also in the virulence phenotype of the resulting mutants: cigR inactivation renders S. Typhimurium hypervirulent (160), whereas mutation of the EIIANtr-encoding ptsN gene slightly attenuates S. Typhimurium virulence (159).
PhoP Promotes Transcription of DNA-Binding Protein SsrB and Vice Versa
PhoP increases transcription of the ssrB gene, which specifies the major regulator of the gene cluster known as Salmonella Pathogenicity Island 2 (SPI-2) (161, 162) (Fig. 4). PhoP promotes ssrB transcription directly, binding to a site located within the upstream gene spiR (also called ssrA), which specifies SsrB’s cognate sensor (77). This control allows transcriptional activation of SPI-2 genes under PhoQ-inducing conditions (77, 161), including those S. Typhimurium experiences inside macrophages (77, 88). In agreement with PhoP promoting ssrB transcription, heterologous expression of SsrB partially restored the requirement of PhoP for SPI-2 gene expression inside macrophages (77). The 5′ leader region of the spiR gene has an inhibitory effect on SpiR amounts, and this inhibition appears to be partially overcome in a PhoP-dependent manner (77). Thus, PhoP activates SsrB both by promoting ssrB transcription and by increasing SpiR abundance.
The SsrB protein, in turn, promotes both PhoP activation and phoP transcription. SsrB is necessary for normal PhoP activation when S. Typhimurium experiences mildly acidic pH and is inside macrophages (88). SsrB binds to the ugtL promoter region and displaces H-NS, increasing ugtL transcription, enhancing PhoQ autophosphorylation (87), and thereby activating PhoP (88) (Fig. 4). Thus, inhibition of SsrB binding to the ugtL promoter region curtails mRNA abundance of PhoP-activated genes while retaining almost wild-type levels of SPI-2 gene expression inside macrophages (88). SsrB also promotes phoP transcription by binding to a site located in the upstream purB gene (88) (Fig. 4). These two actions of SsrB are specific to mildly acidic pH, including within a macrophage phagosome, because SsrB is dispensable for PhoP activation when the PhoQ-inducing signal is low Mg2+ (88). Notably, SsrB impacts transcription of PhoP-activated genes at 6 h after macrophage infection, although initial (albeit not full) PhoP activation takes place at 1 h inside macrophages (88). That SsrB plays a role at later times suggests that SsrB (and/or SpiR) is activated by a phagosomal signal(s)—yet to be identified—different from phagosomal acidification, which takes place within 1 h (82, 83). In sum, the reciprocal activation of PhoP and SsrB allows S. Typhimurium to integrate signals perceived by PhoQ and SpiR/SsrB into PhoP activation, enabling normal transcription of PhoP-activated genes when S. Typhimurium is inside a macrophage phagosome. Thus, acquisition of the spiR and ssrB genes as part of SPI-2 was a critical event in the evolution of S. enterica because it enabled its PhoP/PhoQ system to respond to mildly acidic pH and macrophage signals.
The Physiological Consequences of Altering PhoP-P Amounts under Constant Inducing Conditions
How do the continuous changes in PhoP-P amounts taking place under constant PhoQ-activating conditions benefit an organism? It has been proposed that the surge in PhoP-P allows a bacterium to immediately establish a new phenotypic state (15). For instance, when the PhoQ-activating signal is low environmental Mg2+, this phenotypic state entails changes in the bacterium’s cell surface that decrease its negative charge and thus its dependence on Mg2+ to neutralize the negative charges from the phosphate residues in the various parts of the LPS. The steady-state levels of expression that follow the PhoP-P surge are proposed to maintain the newly established phenotypic state determined by the turnover of the relevant molecules (15).
The PhoP-P protein promotes transcription from the mgtA and mgtC promoters shortly after a shift to low Mg2+ media. However, transcription elongation into the associated coding regions does not proceed until specific cytoplasmic signals are detected by their respective leader mRNAs. This results in the production of small RNAs (sRNAs). In the case of the mgtC leader, the generated sRNA operates in trans as a regulatory sRNA (163). If S. Typhimurium experiences low Mg2+ for an extended period of time and the cytoplasmic Mg2+ concentration decreases below a certain threshold, transcription of the mgtA coding region and of the mgtCBRU-cigR operon takes place. It makes physiological sense for MgtA and MgtB, which import Mg2+ from the periplasm and into the cytoplasm (35), to be produced in response to low cytoplasmic Mg2+ rather than low periplasmic Mg2+ detected by PhoQ. The same is true for MgtC, which decreases the amount of Mg2+-chelating ATP (150, 151).
The positive feedback mechanisms carried out by the MgtA and MgtC proteins increase PhoP-P amount to levels that enable transcription of certain PhoP-activated genes. This regulatory arrangement creates a transcriptional cascade that relies on both a single transcription factor whose abundance changes over time and on promoters that differ in the specific sequences recognized by the PhoP protein as well as in the number, location, and orientation of binding sites for the PhoP-P protein and for other DNA-binding proteins (see below under “PhoP Binding Sites and Promoter Architectures”).
What Signals Activate PhoP when S. Typhimurium Is inside Macrophages
As discussed above, a variety of signals activate the PhoP protein in a PhoQ-dependent manner during bacterial growth in laboratory media. In which environment(s), then, does sensing of the various signals take place? The answer is likely to differ across bacterial species depending on the animal or plant host with which they associate. In addition, the PhoQ protein in a given species may sense different signals in different tissues or even within a given cell type as time progresses and the chemical environment and physical parameters in the bacterium’s surroundings change, often in response to the presence of the bacterium. Here, we discuss PhoP activation in S. Typhimurium and the three best-characterized signals acting on PhoQ: mildly acidic pH, CAMPs, and low Mg2+.
When internalized by mammalian macrophages, the vast majority of S. Typhimurium resides within membrane-bound compartments referred to as Salmonella-containing vacuoles (SCV), which are mildly acidic. Preventing vacuole acidification abolishes activation of the PhoP/PhoQ system (83) and hinders S. Typhimurium replication inside macrophages (82). These results are consistent with the notion that the ability to respond to a mildly acidic pH inside macrophages is essential for activation of PhoP/PhoQ (83), a system that, in turn, is essential for survival inside macrophages (17, 32, 164). Vacuole acidification is required for S. Typhimurium survival in a particular macrophage-like cell line (82), but curiously, not in other macrophages or several epithelial cell lines (165). These results may reflect that bafilomycin A, the pharmacological agent used to prevent phagosome acidification in the experiments just described, may have different effects on different cell types independently of, or in addition to, the inhibition of acidification. Such effects may be responsible for the lack of PhoP/PhoQ activation observed in certain cell types.
A S. Typhimurium strain expressing the PhoQ variant with an artificial cystine bridge between W104C and A128C in the periplasmic domain exhibits wild-type virulence in mice (63, 85). One group found this strain to respond to C18G but not to mildly acidic pH or divalent cations, leading to the proposal that CAMPs are the only infection-relevant signal PhoQ must sense during S. Typhimurium infection of a mammalian host (63). However, PhoP-activated genes were not fully induced when the strain expressing the PhoQ variant with an artificial cystine bridge was inside bone marrow-derived macrophages (63), indicating that PhoQ responds to a signal(s) other than CAMPs. Paradoxically, the same group reported that an S. Typhimurium strain unable to sense C18G because the PhoQ periplasmic domain was replaced by the equivalent region from the P. aeruginosa PhoQ protein retained wild-type virulence (86).
A different group found that the S. Typhimurium strain with the cystine bridge in the periplasmic domain of PhoQ is less responsive to all three signals: C18G, mildly acidic pH, and divalent cations (85). In addition, a S. Typhimurium strain expressing a PhoQ variant with D233E, H409N, and Q460H substitutions in its cytoplasmic domain was attenuated for virulence, albeit not as attenuated as a phoQ-null mutant (85). These three amino acid substitutions reduced virulence even in the strain expressing the cystine-bridged PhoQ, although these amino acid substitutions did not reduce sensing of C18G (85). These results argue that the ability to sense mildly acidic pH is essential for S. Typhimurium virulence in mice (85). Independent support for this notion is provided by the virulence attenuation of a ugtL-null mutant, which is defective in the response to mildly acidic pH but normal in the response to C18G and low Mg2+ (87).
The activity of the artificial peptide C18G in bacteriological media is proposed to reflect that of natural bona fide antimicrobial peptides that S. Typhimurium encounters during infection. However, C18G does not exist in nature (166). Establishing that S. Typhimurium’s ability to respond to C18G is necessary for virulence will require (i) identifying a PhoQ variant specifically defective in sensing C18G (or an equivalent bona fide natural peptide) but retaining a normal response to mildly acidic pH and low Mg2+ and (ii) establishing that an S. Typhimurium engineered to express such a variant instead of the wild-type PhoQ protein is attenuated for virulence. To our knowledge, such a PhoQ variant is yet to be reported.
Intramacrophage Mg2+ concentrations are estimated to be about 1 mM, which is too high a concentration to fully activate PhoQ (18). Moreover, artificially changing the Mg2+ concentration had no effect on PhoP activation inside macrophages (166). The latter measurements were carried out 90 min after S. Typhimurium internalization by macrophages, which is 4 h before the PhoP/PhoQ system is fully active inside macrophages. Nonetheless, ruling out low Mg2+ as a signal detected by Salmonella inside host cells will require evaluating the virulence behavior of strains with PhoQ variants that are not activated in low Mg2+ and yet retain a wild-type response to mildly acidic pH and CAMPs.
Intriguingly, S. Typhimurium experiences Mg2+ starvation in the spleens of mice harboring a functional Slc11a1 protein, as manifested by the specific induction of the PhoP-activated Mg2+ transporter MgtB (167). Slc11a1 (previously referred to as Nramp1) is a host defense protein that localizes to the macrophage phagosomal membrane and confers resistance to a variety of intracellular pathogens that remain within a membrane-bound vacuole of mildly acidic pH (168). Moreover, an mgtB mutant is defective for survival inside Slc11a1+/+ macrophages but normal in Slc11a1–/– macrophages (169) and fails to reach a wild-type optical density after extended incubation in laboratory media of low Mg2+ (169).
Critically, bacterial species that harbor functional PhoP/PhoQ systems differ in the content of horizontally acquired genes, some of which control the activity of the ancestral PhoP/PhoQ system in response to specific signals. As discussed above, PhoQ activation in mildly acidic pH requires the horizontally acquired ugtL (87) and ssrB (88) genes in S. Typhimurium (Fig. 3A) and the horizontally acquired safA gene in E. coli (90, 93) (Fig. 3B). These genes specify proteins with little significant amino acid sequence similarity. Therefore, it is not possible to exclude the possibility of a signal activating PhoQ solely because a particular gene(s) is absent from a given genome. In addition, allelic differences among conserved genes can give rise to distinct phenotypic behaviors in related species (170,–173).
To conclude, PhoQ responds to changes in cytoplasmic pH when inside macrophages, enabling S. Typhimurium to integrate extracytoplasmic host-derived signals and bacterial cytoplasmic signals into the activation status of its cognate regulator PhoP (174).
THE REGULATORY ACTIVITY OF PhoP: MULTIPLE WAYS TO CONTROL GENE EXPRESSION
PhoP controls the abundance of a large number of proteins in S. Typhimurium (175). These proteins can be divided into two general groups based on whether they are controlled directly by PhoP at the transcriptional level or indirectly by PhoP-regulated DNA-binding transcriptional regulators, anti-silencers, or proteins that alter the stability of other proteins or the abundance of metabolites (Fig. 5). PhoP also controls transcription of genes specifying regulatory RNAs, some of which regulate S. Typhimurium virulence in animal hosts (176, 177) (Fig. 5). In this section, we discuss how PhoP directly alters transcription of target genes by binding to specific DNA sequences and the mechanisms by which PhoP-regulated gene products alter protein abundance.
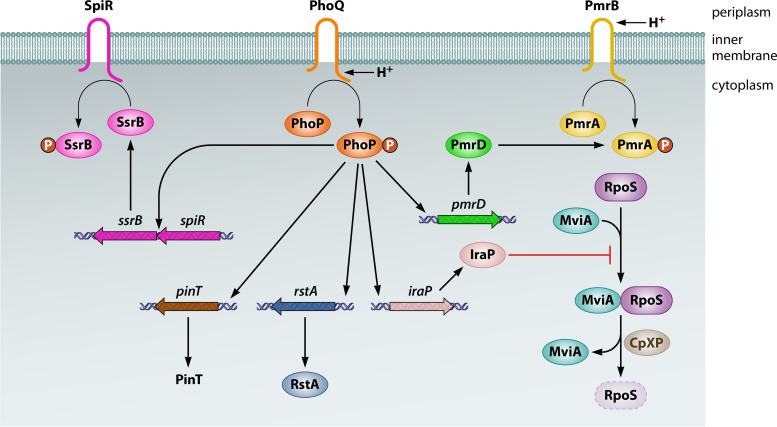
Activation of regulatory proteins and RNAs by PhoP in S. Typhimurium. PhoP is a direct transcriptional activator of the ssrB and rstA genes, which specify DNA-binding regulatory proteins, and of the pinT gene, which specifies a regulatory RNA. PhoP activates the regulator PmrA by promoting transcription of pmrD, which specifies a protein that hinders PmrA-P dephosphorylation by the sensor PmrB. PhoP prevents degradation of the alternative sigma factor RpoS by promoting transcription of iraP, which specifies a protein that binds to the protease adaptor MviA, thereby preventing MviA from binding RpoS and thus sparing RpoS proteolysis by ClpXP. Whereas E. coli has a highly conserved IraP protein, its iraP gene is not transcriptionally activated by PhoP, which promotes transcription of a different anti-adaptor encoding gene named iraM.
How PhoP Promotes Transcription of PhoP-Activated Genes
PhoP consists of an N-terminal 124-amino-acid domain that exhibits sequence similarity to response regulators of the two-component system family (118) and a C-terminal 99-amino-acid domain that harbors the winged helix-turn-helix motif responsible for DNA binding. This organization is characteristic of members of the OmpR/PhoB subfamily of response regulators, to which PhoP belongs (178). Crystal structures for the N-terminal domain of the E. coli PhoP protein in its unphosphorylated form and in the presence of the phosphoryl analog beryllofluoride showed that both the inactive and active forms of this PhoP domain dimerize with a 2-fold symmetry (179), resembling the behavior of dimers of the activated form of the related domains from the OmpR and PhoB proteins.
PhoP phosphorylation stabilizes the active conformation of PhoP’s regulatory domain (179). The crystal structure of PhoP’s receiver domain obtained in the absence of the activating agent beryllofluoride resembled that of the active structure (179). This similarity may be the result of the high PhoP concentration used during crystallization. Gel filtration studies are consistent with inactive PhoP being a monomer and active PhoP a dimer (179). That PhoP recognizes an imperfect tandem repeat (see “PhoP Binding Sites and Promoter Architectures”) and that PhoP’s regulatory domain exhibits a 2-fold symmetry support the proposal that the active form of the PhoP protein is a dimer in which regulatory and DNA-binding domains are joined by flexible linkers (179), which assumes a head-to-tail orientation of the DNA-binding domains.
In vitro experiments using His-tagged PhoP led to the conclusion that PhoP phosphorylation enhances PhoP dimerization, which favors DNA binding (55). The conformation of the purified His-tagged PhoP protein changed upon autophosphorylation from acetyl phosphate, rendering the C-terminal DNA-binding domain resistant to trypsin (180). In agreement with these in vitro results, the ability of PhoP to be phosphorylated at the conserved aspartate residue (i.e., D52) is necessary for DNA binding by a C-terminally hemagglutinin (HA)-tagged PhoP protein in vivo (14). The transcriptional surge and virulence properties of strains expressing PhoP-HA and untagged PhoP are similar (14, 15). However, others found that C-terminally His-tagged PhoP-P and untagged PhoP-P differ in their in vitro behaviors; that is, phosphorylation did not affect dimerization or DNA binding of untagged PhoP but promoted both properties in His-tagged PhoP (181). These results suggest that wild-type PhoP exists in equilibrium between its monomeric and dimeric forms and that a C-terminal His tag alters the biochemical properties of PhoP in vitro. Comparison of the in vivo behavior of isogenic strains expressing His-tagged and untagged PhoP proteins may establish whether the His tag alters the properties of the PhoP protein in living cells.
PhoP Binding Sites and Promoter Architectures
PhoP-P recognizes and binds a specific, degenerate DNA sequence, designated the PhoP box. Promoters directly activated by PhoP-P differ not only in the specific nucleotides that constitute the PhoP box but also in the number of PhoP boxes, the distance between the PhoP box(es) and the −10 region, and the orientation of the PhoP box(es) relative to promoter sequences, which altogether define the promoter architecture (Fig. 6). The architectures that PhoP-P actually uses in vivo have specific combinations of the promoter elements just described.

Promoter architectural landscape of the S. Typhimurium PhoP regulon. Architecture and DNA sequences of PhoP‐activated promoter regions are shown. DNA sequences corresponding to 23 such promoters are aligned with respect to the predicted −10 hexamer using the 5′-most edge of this element as a referential landmark instead of the typically used transcription start site (indicated in green). PhoP box orientation is indicated with blue arrows. The predicted −35 hexamer is underlined in red, and the predicted −10 hexamer is indicated in red. PhoP boxes sharing common patterns, including orientation, phasing, and/or location with respect to the RNAP, are indicated with grey boxes. The promoters are divided into five classes based on their shared cis‐acting features. From top to bottom: architecture I, architecture II, architecture III, architecture IV, and architecture V. (Reproduced from reference 10 with permission of the publisher.)
A single PhoP box overlaps the −35 element in canonical PhoP-activated promoters, such as those driving transcription of the phoP and mgtA genes (10) (Fig. 6). This location suggests that PhoP utilizes a class II activation mechanism (182) at the corresponding promoters (183,–185). Class II activation entails interactions between an activator protein and subunits of RNA polymerase other than the C-terminal domain of the α subunit (α-CTD) (182). In agreement with this notion, PhoP activation of the mgtA promoter in vitro does not require the α-CTD (180). PhoP-P binding to the PhoP box in the mgtA promoter is stimulated by RNA polymerase and, in turn, RNA polymerase binding to the mgtA promoter is stimulated by PhoP-P (180).
In other promoters, the PhoP box is located further upstream (up to 78 nt upstream of the transcription start site) (10), suggesting class I activation (i.e., binding to α-CTD [182]) (Fig. 6). Accordingly, rstA activation requires the α-CTD of RNA polymerase (10). Additional PhoP-activated promoters in this category are those driving transcription of the mgtC, pagC, and ugtL genes.
Initial work aligning a limited set of PhoP-regulated promoter regions identified the direct repeat (G/T)GTTTA separated by 5 nt as the PhoP box (13). This motif was later refined and led to the identification of PhoP box submotifs. In addition to the canonical (G/T)GTTTA repeat termed S1, PhoP-P also binds the asymmetrical (T/C)ATTTA—5 nt—(G/T)GTTTA (S2) and (G/T)GTTTA—5
nt—(G/T)GTTTA (S2) and (G/T)GTTTA—5 nt—TATTTA (S3) sites (10). The diversity of PhoP box motifs has the potential of conferring distinct properties on PhoP-regulated promoters, such as being controlled by other DNA-binding proteins that recognize sequences that overlap the PhoP box.
nt—TATTTA (S3) sites (10). The diversity of PhoP box motifs has the potential of conferring distinct properties on PhoP-regulated promoters, such as being controlled by other DNA-binding proteins that recognize sequences that overlap the PhoP box.
Although 12 different PhoP box submotifs have been identified (186), five of them are primarily used by PhoP to promote transcription in S. Typhimurium (10). The architectures of PhoP-activated promoters (i.e., the combination of PhoP box motif, number, location, and orientation) differ between those driving transcription of ancestral genes and those controlling horizontally acquired genes and do not reflect the function of the corresponding gene products (10) (Fig. 6). That is, promoters with a single PhoP box tend to control ancestral genes, whereas those with two or more PhoP boxes tend to drive transcription of horizontally acquired genes (10). For instance, the promoter driving transcription of the ancestral mgtA gene has a different architecture from that activating transcription of the horizontally acquired mgtB gene (10) (Fig. 6), even though the mgtA and mgtB genes specify Mg2+ transporters that share 50% amino acid identity (35).
Some promoters harbor the PhoP box in the opposite orientation of that found in the canonical mgtA and phoP promoters (10) (Fig. 6). The distance from the PhoP box to the RNA polymerase binding site corresponds to multiples of 10 to 11 nucleotides (i.e., one turn of a DNA helix), suggesting that these promoters share the phase in which the PhoP protein is present. The distance between the PhoP box and the −10 region and the orientation of the PhoP box ultimately dictate the face of the DNA helix in which the PhoP-P protein binds and thus its ability to establish productive contacts with RNA polymerase.
Critically, the architecture of certain PhoP-activated promoters is species specific. For example, the particular combination of PhoP box motif, number, location, and orientation found in the S. Typhimurium mgtC promoter has not been reported in Y. pestis, which harbors a functional PhoP/PhoQ system (46). Likewise, the combination of PhoP box motif, number, location, and orientation found in the Y. pestis mgtC promoter has not been found in S. Typhimurium (46). One of the consequences of the species-specific nature of promoter architectures is that the S. Typhimurium PhoP protein is unable to activate transcription of the Y. pestis mgtC gene, and that the Y. pestis PhoP protein is unable to activate transcription of the S. Typhimurium mgtC gene (46). This is in spite of the fact that both PhoP proteins bind equally well to both mgtC promoters (46). These results reflect that the genes specifying DNA-binding regulatory proteins coevolve with the promoter sequences targeted by those proteins. Therefore, horizontal acquisition of a gene harboring a PhoP box in its promoter region does not guarantee PhoP-regulated expression of that gene in a different species.
The different architectures of PhoP-activated promoters enable bacteria to exert distinct control over different members of a regulon within a given species. This is of special importance given PhoP’s various roles in virulence and Mg2+ homeostasis (35). The differential expression of PhoP-activated gene products is also achieved by unusually long 5′ leader regions (10) that control whether the associated coding regions are expressed in response to metabolites, sRNAs, proteins, or the ribosome.
PhoP Overcomes Gene Silencing by the H-NS Protein in Multiple Ways
H-NS is a nucleoid-associated protein that binds to a degenerate AT-rich sequence and then extends over neighboring regions, silencing expression of the corresponding genes (187). Because horizontally transferred genes tend to be AT-rich, H-NS is often referred to as a xenogeneic silencer (188). PhoP uses three different mechanisms to overcome gene silencing by H-NS. First, the PhoP box is AT-rich, enabling PhoP-P to outcompete H-NS for binding at certain AT-rich sequences. Such activity may suffice to derepress a gene or operon or constitute the first step toward gene expression, as RNA polymerase recruitment may also require PhoP-P or a different DNA-binding protein once a promoter is cleared of H-NS.
Second, PhoP promotes transcription of genes specifying DNA-binding proteins with the ability to overcome gene silencing by H-NS. For example, transcription of the horizontally acquired H-NS-silenced genes ugtL and pagC requires the PhoP-activated anti-silencing protein SlyA (7) in addition to PhoP (189). SlyA displaces H-NS from the ugtL and pagC promoter regions but is unable to activate gene transcription by itself. PhoP activates transcription from these two promoters once H-NS has been displaced by SlyA (189).
It has been suggested that PhoP-activated promoters can be divided into three groups based on their architectural features and the role played by the PhoP-P, SlyA, and H-NS proteins at such promoters (190). Because hns is an essential gene in S. Typhimurium (191, 192), PhoP-dependent gene transcription was examined in a set of strains with a hypomorphic allele of the rpoS gene that confers tolerance to hns inactivation (190). (hns inactivation is also viable in a phoP mutant [192].) The authors of that study argued that PhoP-activated promoters with a single PhoP box drive transcription of ancestral genes (190) and are not subject to silencing by H-NS (190). By contrast, PhoP-activated promoters with more than one PhoP box correspond to those activating H-NS-silenced horizontally acquired genes (190). However, there are a single PhoP box in the promoter of the horizontally acquired orgB gene and two PhoP boxes in the promoter of the ancestral pagP gene (10), indicating that PhoP box number at a promoter is not necessarily a predictor of gene ancestry.
In vitro transcription experiments with supercoiled DNA templates support the notion that the PhoP and SlyA proteins activate transcription of promoters with more than one PhoP box by overcoming silencing by H-NS rather than by recruiting RNA polymerase (190). These results are in contrast to those obtained with linear DNA templates demonstrating that SlyA is necessary solely to overcome H-NS silencing at the pagC and ugtL promoters, whereas PhoP-P is necessary to overcome silencing and to recruit RNA polymerase (189).
The Shi group found H-NS binding to and repression of the S. Typhimurium phoP promoter (193), but other groups did not detect such effects (191, 192). The use of an hns-null mutation by the Shi group (194) raises the possibility of the tested strains having an additional mutation(s) because an hns null mutation is lethal (191, 192). The Shi group also reported that certain divergently transcribed genes harbor similarly located binding sites for the PhoP and SlyA proteins in the corresponding intergenic regions (195) and that PhoP and SlyA overcome silencing of such gene pairs by H-NS (195). However, the proposed regulation has been validated only for a few genes.
The PhoQ-activating condition low Mg2+ promotes expression of the H-NS-silenced sRNA Stnc1480 even though the H-NS protein remains bound to the regulatory region of this gene (196). SlyA promotes Stnc1480 expression by overcoming H-NS silencing because Stnc1480 expression was restored to the slyA mutant upon hns inactivation (196). By contrast, no expression was observed in a phoP mutant despite the absence of hns (196), arguing that PhoP is necessary to recruit RNA polymerase to the Stnc1480 promoter (196). This behavior is reminiscent of that displayed by the PhoP and SlyA proteins at the pagC promoter (189). A situation in which PhoP itself displaces H-NS from grhD1 has also been proposed (197), although SlyA involvement has not been investigated, and direct competition between PhoP and H-NS has not been reported.
The identification of multiple SlyA-dependent, PhoP-activated horizontally transferred genes (26) suggests that these H-NS-silenced genes are activated by mechanisms resembling those acting on the pagC and ugtL promoters (discussed above). The requirement of two proteins for full expression of H-NS-silenced genes contrasts with the anti-silencing mechanisms mediated by the LeuO and ToxT proteins, by which binding of a single anti-silencer protein to a promoter region displaces H-NS or prevents filament formation and is sufficient to promote gene transcription (198).
In addition to SlyA, the PhoP-activated anti-silencing protein SsrB (77, 199) enables transcription of some H-NS silenced genes. For example, transcriptional activation of the H-NS-silenced ugtL gene (192) in mildly acidic conditions requires SsrB to antagonize H-NS (88). Unlike SlyA (189), however, SsrB is dispensable for ugtL transcription in low Mg2+ (88). Despite the common action of SlyA and SsrB (i.e., displacing H-NS), these proteins bind distinct locations of the ugtL promoter region (88, 189). Thus, PhoP requires different DNA-binding proteins to transcribe the ugtL gene, depending on the identity of the PhoQ-inducing signal, which may or may not activate other regulators. Furthermore, PhoP promotes expression of H-NS-silenced SsrB-activated genes located within and outside the SPI-2 locus by increasing transcription of the ssrB gene (77, 161, 192, 200,–202).
Finally, the third mechanism by which PhoP overcomes gene silencing is by decreasing H-NS amounts, thereby promoting expression of H-NS-silenced genes genome wide. PhoP facilitates H-NS degradation when S. Typhimurium experiences mildly acidic pH or is present inside a macrophage phagosome (203). When bound to DNA, H-NS is protected from the protease Lon (203). However, PhoP renders H-NS vulnerable to Lon by displacing H-NS from DNA sites throughout the S. Typhimurium genome and, to a much lesser extent, by transcription-induced detachment of H-NS from PhoP-activated promoter regions (203). The PhoP-activated SsrB (77) and SlyA (7) proteins are necessary for full derepression of H-NS-silenced genes (203). By promoting H-NS degradation, S. Typhimurium derepresses H-NS-silenced genes without having to evolve a PhoP box(es) at each H-NS-silenced gene. Which H-NS-silenced gene(s) is derepressed upon H-NS degradation is determined by the affinity of H-NS for its target DNAs, with tight binders being impervious to PhoP action (203). Despite activating PhoP as much as mildly acidic pH, growth in low Mg2+ does not result in H-NS proteolysis in S. Typhimurium (203). Wild-type S. Typhimurium decreases H-NS amounts 95% by 6 h inside macrophages (203), suggesting that H-NS proteolysis is the dominant mechanism of derepression of horizontally acquired H-NS-silenced genes in this environment.
PhoP as a Transcriptional Repressor
The vast majority of PhoP-regulated genes reported to date are PhoP activated. However, PhoP can operate (i) as a transcriptional repressor at certain promoters, (ii) as an activator of one promoter while serving as repressor of the promoter for a divergently transcribed gene, or (iii) as an activator and repressor at the same promoter, depending on growth conditions.
PhoP represses transcription of the clpS gene by binding to a region of the clpS promoter that overlaps the −10 element and the transcription start site (16). This repression requires the mgtA- and mgtC-dependent increase in active PhoP amounts (16). That the dependence on mgtC is stronger than on mgtA (16) is the opposite of what is observed in PhoP-activated genes, such as pagC, that exhibit a stronger dependence on mgtA than on mgtC (57, 58). Although the clpS gene is immediately followed (i.e., 31 nt downstream) by the similarly oriented clpA gene, PhoP-activating conditions reduce clpS expression but do not alter that of clpA. The specific control that PhoP exerts over ClpS abundance enables bacteria to establish a hierarchy among ClpS-dependent substrates of the ClpAP protease with minimal effects on ClpS-independent ClpAP substrates (16).
PhoP binding to the mgtA-treR intergenic region promotes transcription of the mgtA gene while repressing transcription of the divergently transcribed treR gene (180). TreR is a transcriptional repressor of the treBC genes mediating trehalose utilization. Although α-CTD is dispensable for mgtA activation by PhoP, it is necessary for repression of treR (180). The physiological reason for PhoP simultaneously promoting mgtA expression and treR repression, though currently unknown, may be part of the bacterial response to osmolarity changes because (i) hyperosmolarity activates the PhoP/PhoQ system (96), (ii) hyperosmotic stress favors transcription elongation into the mgtA coding region (99), and (iii) trehalose is an osmoprotectant.
Several PhoP-activated promoters with two PhoP boxes harbor one at a position normally associated with promoter repression, such as overlapping the −10 element or downstream of the transcription start site, and the other located upstream, at a position associated with promoter activation (10). Because PhoP-P binds with stronger affinity to the upstream than downstream PhoP box, such promoters may first be activated by PhoP-P and then shut off once PhoP-P reaches a certain concentration. For example, one of the PhoP boxes in the ugtL promoter overlaps the −35 region, and the other one overlaps the −10 region (7). Mutations in the −35-region-overlapping PhoP box that prevent PhoP binding abolish ugtL transcription. The role of the −10-region-overlapping PhoP box awaits the test of mutants that abolish PhoP binding without compromising RNA polymerase recognition of the −10 region.
Two-dimensional gel electrophoresis of protein extracts from isogenic wild-type and pho-24 S. Typhimurium revealed that PhoP decreases the amounts of several proteins (204). A complementary genetic approach discovered a PhoP-repressed gene mapping to Salmonella pathogenicity island 1 (SPI-1) and mediating invasion of epithelial cells (205). We now know that PhoP decreases expression of the type III secretion apparatus specified in SPI-1 by repressing transcription of the hilA gene (206) and promoting transcription of the sRNA PinT (207). HilA is the master activator of SPI-1 genes (208). PhoP represses hilA expression directly by binding to its promoter region and indirectly by repressing transcription of the hilA activator genes hilD and rtsA that, in turn, antagonize the silencing effects that H-NS exerts on the hilA promoter region (206). The PhoP-activated PinT, in turn, represses translation of both hilA and rtsA (207).
In terms of S. Typhimurium’s course of infection in an animal host, PhoP represses transcription of genes involved in bacterial entry into epithelial cells but activates those that aid replication and survival inside phagocytic cells.
Direct Transcriptional Activation of DNA-Binding Proteins
In addition to the direct regulation discussed above, PhoP controls gene expression indirectly, by a variety of mechanisms presented in the following sections. Here, we discuss how PhoP promotes transcription of genes specifying DNA binding transcriptional regulators that, in turn, alter transcription of other genes (Fig. 5).
PhoP binds to the promoter of the ssrB gene and is required for ssrB transcriptional induction when S. Typhimurium is inside macrophages (77). The SsrB protein is predicted to form a two-component system with the sensor SpiR (162), required for S. Typhimurium virulence (209, 210). SsrB promotes expression of a type III secretion system and a variety of effectors translocated by this system into the host cell and required for the intracellular lifestyle of S. Typhimurium (211). Although SsrB/SpiR, the type III secretion system, and secreted effectors are encoded within the horizontally acquired SPI-2 (200), SsrB also promotes transcription of genes located throughout the S. Typhimurium genome (200) and represses genes in SPI-1 (212). Therefore, a phoP mutant displays decreased abundance of the translocated proteins SseK1 and SseK2 (213) and SopD and SopD2 (202) because they are encoded by genes transcriptionally controlled by SsrB.
The vast majority of the time, the genes specifying a given two-component system are part of the same operon (128). However, spiR transcription is phoP independent under conditions in which ssrB transcription is phoP dependent (77). This is in contrast to the transcriptional control of both spiR and ssrB exerted by OmpR (214). Nonetheless, PhoP appears to activate spiR expression posttranscriptionally, which is believed to happen via the 5′ leader region of the spiR transcript (77). OmpR and PhoP control ssrB expression at early and late times, respectively, following S. Typhimurium internalization by murine macrophages (77, 78).
The PhoP-activated ssrB promoter is directly repressed by the PmrA protein (215). This is paradoxical because PhoP activates PmrA (40) (see below under “Posttranslational Control of DNA-Binding Regulatory Proteins and Unusual PhoP Role at Certain Promoters”). This means that PhoP both promotes ssrB transcription directly and enhances the active state of a protein that represses ssrB transcription directly (Fig. 5). Binding of the PmrA protein to the ssrB promoter region does not hinder PhoP binding to that promoter and vice versa (215). Inactivation of the pmrA gene or the PmrA binding site in the ssrB promoter increases S. Typhimurium virulence in C3H/HeN mice (215), which are Slc11a1+/+. (A group reported that a S. Typhimurium pmrA-null mutant is mildly attenuated in C57BL/6 mice, which are Slc11a1–/–, when inoculated intraperitoneally with an unusually high dose corresponding to 100,000 ×
× the median lethal dose [LD50] [216].) The PhoP and PmrA proteins likely control ssrB transcription at different times when S. Typhimurium is inside macrophages.
the median lethal dose [LD50] [216].) The PhoP and PmrA proteins likely control ssrB transcription at different times when S. Typhimurium is inside macrophages.
The PhoP control of ssrB expression provides a singular example of a transcription factor—PhoP—that, despite being essential for virulence (17, 31, 32), increases the abundance of both virulence (i.e., SsrB) and antivirulence (i.e., PmrA) regulatory proteins. Antivirulence proteins are those that, upon inactivation, increase the virulence of a pathogen.
PhoP is a direct transcriptional activator of the slyA gene (7), which, as discussed above, specifies a DNA-binding protein that antagonizes H-NS silencing at some PhoP-activated promoters (7, 26, 189). Although guanosine pentaphosphate/tetraphosphate [(p)ppGpp] was proposed to be an activating ligand for the S. Typhimurium SlyA protein (195), these findings have not been recapitulated with the highly conserved E. coli SlyA protein (217). Moreover, SlyA crystal structures (218, 219) make it unlikely that SlyA binds (p)ppGpp.
The RstA/RstB two-component system regulates pyrimidine metabolism, iron acquisition, biofilm formation, and acid resistance (220, 221). A proteomic analysis showed a partial overlap between the RstA/RstB and PhoP/PhoQ regulons of S. Typhimurium (220). The rstA gene is followed by the rstB gene in the E. coli genome but separated from rstB by two genes in the S. Typhimurium genome. RstA and RstB do not appear to function as a two-component system in S. Typhimurium because rstB was dispensable for RstA-promoted transcription of the feoB gene (222).
The PhoP/PhoQ system is necessary for rstA expression in both S. Typhimurium and E. coli (222, 223) (Fig. 5). PhoP binds to the rstA regulatory region, indicating direct rstA activation (10, 221). This binding, however, is insufficient to promote transcription of RstA-activated genes in S. Typhimurium because RstA needs to be phosphorylated by a currently unknown phosphoryl donor that operates at a mildly acidic pH but not in low Mg2+ (222). Thus, RstA-activated genes are expressed only in mildly acidic pH, even though PhoP promotes rstA transcription when the PhoQ-activating signal is mildly acidic pH or low Mg2+ (224).
When expressed from a multi-copy-number plasmid, the rstA gene increases iron uptake by the FeoB protein encoded in the feoAB operon (224). The imported iron forms a complex with the Fur protein that represses expression of genes mediating iron uptake, metabolism, and storage (224). The connection between the PhoP/PhoQ system and iron homeostasis is reinforced by the hypersusceptibility of a phoP-null mutant toward Fe(II) (225).
PhoP-Activated Regulatory RNAs
PhoP controls the abundance of at least 33 sRNAs in S. Typhimurium: 25 activated and 8 repressed (196). PhoP regulates some of these RNAs directly and others indirectly, by controlling the abundance of other regulatory proteins (Fig. 5). The vast majority of the sRNAs indirectly controlled by PhoP/PhoQ are also regulated by SsrB/SpiR, possibly reflecting that this regulation was discovered using inducing conditions for the SsrB/SpiR system (196), which also activate PhoP/PhoQ but not to the full extent.
The sRNA Stnc1480 is specified by an H-NS-silenced gene that requires both PhoP and SlyA, but not the OmpR/EnvZ and SsrB/SpiR systems, for expression (196). SlyA operates as an antisilencer at the stnc1480 promoter, whereas PhoP is still required to activate stnc1480 transcription when hns is absent. The target(s) of Stnc1480 remains unknown.
The 219-bp intergenic region that separates the mgtC stop codon from the mgtB start codon in the mgtCBRU-cigR operon harbors a PhoP-activated promoter that drives transcription of a 1.2-kb antisense RNA complementary to the mgtC portion of the sense mgtCBRU-cigR transcript (176). Termed AmgR, this regulatory RNA destabilizes the mgtC portion of the transcript in an RNase E-dependent but RNase III- and Hfq-independent manner (176). Even though AmgR does not exhibit complementarity to the mgtB part of the transcript, AmgR expression lowers not only MgtC but also MgtB amounts, albeit to a lesser extent the latter (176).
PhoP-P plays an unusual role in regulating expression of the MgtC and MgtB proteins because it activates the promoters of both: the sense RNA for the mgtCBRU-cigR operon (13, 153, 154) and the antisense AmgR (176) (Fig. 5). Given that PhoP-P has higher affinity for the mgtC promoter than for the amgR promoter, MgtC and MgtB synthesis proceeds over a time window following exposure to inducing conditions until PhoP-P activates the amgR promoter. Inactivation of the amgR promoter renders S. Typhimurium hypervirulent (176), whereas mgtC inactivation attenuates virulence (158). This provides yet another example of PhoP activating transcription of an antivirulence gene (amgR) that decreases expression of PhoP-activated virulence genes (mgtC and mgtB).
The first 113 nt of the 296-nt leader RNA that precede the mgtC coding region constitute a trans-acting sRNA that reduces expression of both FljB, one of the two flagellar types produced by biphasic Salmonella, and FljA, a repressor of FliC, the other flagellar type (163). This PhoP-activated sRNA promotes degradation of the fljB/fljA transcript in a manner dependent on Hfq and RNase E (163). Although this sRNA includes the ORF mgtM, translation of which controls transcription elongation into the associated coding region (100, 156), replacing mgtM’s start codon by a stop codon did not compromise the ability of the sRNA to repress fljB expression (163). Therefore, transcription activation of the mgtC promoter generates a leader RNA that can act in trans to downregulate motility and in cis to control transcription elongation into the coding region of the mgtCBRU-cigR operon. It is presently unknown what condition(s) favors the production of the 113 nt sRNA rather than longer versions of the leader region of the mgtCBRU-cigR transcript, as termination factor Rho promotes transcription termination downstream of position 113 (226).
PinT is a PhoP-activated sRNA highly expressed when S. Typhimurium is inside a variety of mammalian cells (177) (Fig. 5). The Salmonella-specific 80-nt PinT has multiple targets: it represses expression of the sopE and sopE2 genes, which specify effectors with a role in bacterial invasion of epithelial cells (227). PinT activity is Hfq dependent and proposed to serve as a timer of virulence gene expression (177). PinT inhibits translation of the gene specifying the secreted effector SteC (228) and, as discussed above, of the invasion regulatory genes hilA and rtsA (207).
MgrR is a PhoP-activated sRNA that represses expression of the eptB gene in both E. coli and S. Typhimurium (59, 229). The eptB gene product mediates the covalent modification of the outer 3-deoxy-d-manno-octulosonic acid in the core region of the LPS with phosphoethanolamine (230). When mgrR is absent, eptB expression increases, which enhances this LPS modification (231) and results in a modest increase in resistance to the cationic peptide antibiotic polymyxin B (59). Because the mgrR promoter has a conserved extended −10 region, it requires only moderate amounts of active PhoP protein for transcription, which is reflected by a phoQ-null mutant exhibiting wild-type mgrR expression (59). The properties of the mgrR promoter allow mildly inducing conditions for PhoQ to promote mgrR transcription (59). The mgrR expression behavior differs from that of the vast majority of PhoP-activated genes whose expression is equally abolished in phoP and phoQ mutants (18, 123). For instance, mgtS (also referred to as yneM) is a gene convergently transcribed with mgrR that requires both phoP and phoQ for expression in E. coli (59). The downregulation of EptB carried out by MgrR is in contrast to the PhoP-dependent upregulation of other genes specifying phosphoethanolamine transferases that modify other LPS sites and/or render bacteria resistant to polymyxin B (36, 139, 232).
Transcription of the eptB gene is also promoted by RNA polymerase programmed with sigma E (231), which is liberated from the inner membrane when bacteria experience specific outer membrane disruptions (233). As stated above, sigma E is also responsible for driving transcription of the sRNA MicA, a repressor of phoP translation (119). Therefore, sigma E and PhoP have opposite effects on eptB expression, which results in the incorporation of phosphoethanolamine at different positions of the LPS, thereby altering the properties of the outer membrane.
Posttranslational Control of DNA-Binding Regulatory Proteins and Unusual PhoP Role at Certain Promoters
In addition to its direct transcriptional effects on regulatory genes, PhoP-P alters the abundance or activity of DNA-binding regulatory proteins indirectly, via PhoP-regulated gene products. For instance, the PhoP-activated pmrD gene specifies a protein that binds to PmrA-P, preventing its dephosphorylation by PmrB when S. Typhimurium does not experience activating conditions for PmrB (40) (Fig. 5). PmrD expands the spectrum of environments in which PmrA-activated genes are expressed by including those activating the noncognate sensor PhoQ (e.g., low Mg2+) (41). PhoP/PhoQ’s role in activating PmrA is solely to promote PmrD expression because pmrD transcription from a heterologous promoter results in transcription of PmrA-activated genes in phoP- and phoQ-null mutants (41).
The PmrA protein, in turn, is a transcriptional repressor of the pmrD promoter (6). This repression prevents accumulation of unhealthy PmrA-P amounts in bacteria experiencing PmrB-activating signals. PmrA activation by its cognate sensor PmrB is pmrD independent in S. Typhimurium (41). By contrast, E. coli requires pmrD to activate PmrA when experiencing PmrB-activating conditions (234, 235). However, E. coli is unable to activate PmrA under PhoQ-activating conditions (173) or to repress the pmrD promoter by PmrA (173). The E. coli pmrD gene specifies a protein that is only 55% identical to the S. Typhimurium PmrD (173), and the E. coli PmrB has a more potent phosphatase activity toward PmrA-P than the S. Typhimurium PmrB (170). As discussed below (see “Evolution of PhoP and Its Regulon”), the PhoP-PmrD-PmrA activation pathway is also present in K. pneumoniae (236).
The PmrA-activated ugd gene specifies UDP-glucose dehydrogenase, an enzyme that converts UDP-glucose into UDP-glucuronic acid (127). UDP-glucuronic acid is a precursor in the synthesis of both colanic acid capsule and 4-aminoarabinose, a chemical modification of the lipid A phosphates that reduces the overall negative charge of the LPS and renders the bacterium resistant to polymyxin B (127). The ugd gene is also activated by RcsB (79), a transcriptional regulator that controls expression of other colanic acid synthesis genes when forming a heterodimer with the RcsA protein (237). Although PhoP binds to a DNA region upstream of the ugd promoter (80), the ugd gene can be expressed in a phoP-independent manner under different laboratory conditions (79). This expression is mediated by a PmrA homodimer, an RcsB homodimer, or an RcsB-RcsA heterodimer, depending on the particular stress condition experienced by S. Typhimurium. There is no ugd expression in a pmrA rcsB double mutant, indicating that PhoP is unable to promote ugd transcription on its own. By contrast, ugd expression is strictly dependent on phoP when S. Typhimurium is inside macrophages (79). Therefore, PhoP promotes ugd expression in two ways: (i) by activating transcription of the pmrD gene, which activates the PmrA protein posttranslationally (40) and results in PmrA binding to the ugd promoter, and (ii) by working together with the RcsB protein in ways yet to be determined (80).
PhoP Controls Protease Activity and Specificity
E. coli and S. Typhimurium harbor five cytosolic ATP-dependent proteases, designated ClpAP, ClpXP, FtsH, Lon, and HslVU (238). These proteases use the energy derived from ATP hydrolysis to unfold their substrates, which are then shuttled to a proteolytic chamber or protease subunit for degradation (239). The PhoP-activated gene products MgtA, MgtB, and MgtC reduce ATP abundance (38), and changes in ATP abundance are correlated with corresponding changes in the abundance of the ClpAP substrate PhoP (58) and ClpXP substrate RpoS (240). The activities of the Lon and FtsH proteases toward their respective substrates is also impacted by the PhoP-dependent decrease in ATP abundance (HslVU substrates were not investigated) (37).
In addition to the systemic effects on proteolysis resulting from changes in ATP amounts (37), PhoP controls transcription of products that modify the degradation of specific protease substrates. As discussed above, PhoP is a transcriptional repressor of the ClpAP adaptor ClpS (16) (Fig. 5). Therefore, under PhoP-activating conditions, ClpS abundance decreases 8-fold, which reduces proteolysis of a subset of ClpS-dependent substrates of ClpAP (16), including the putrescine amino transferase Oat and the transcriptional regulator SirA (UvrY in E. coli). By contrast, the stability of those ClpS-dependent substrates that bind ClpS strongly, such as FtsA, remains unaltered (16). The PhoP-mediated repression of clpS expression results in Oat-dependent antibiotic resistance and SirA-dependent gene transcription (16). The latter control may have genome-wide effects because SirA, which regulates virulence and biofilm formation (241), is a transcriptional activator of the sRNA genes csrB and csrC. The CsrB and CsrC sRNAs interact with the RNA binding protein CsrA, a pleiotropic regulator of gene expression that binds to sequences that resemble a ribosome binding site (242). The synthase/hydrolase of (p)ppGpp SpoT is also a ClpS-dependent ClpAP substrate. (p)ppGpp controls expression of hundreds of genes due to its interactions with RNA polymerase (243) and the biochemical activities of different enzymes (244).
PhoP promotes transcription of two different anti-adaptors that prevent the protease adaptor MviA from delivering RpoS to ClpXP for degradation (152). In S. Typhimurium, PhoP promotes transcription of the anti-adaptor gene iraP, which stabilizes RpoS, enabling reprogramming of RNA polymerase with RpoS to transcribe RpoS-dependent promoters in response to PhoQ-inducing signals (Fig. 5). Growth under PhoQ-activating conditions results in H2O2 resistance by the RpoS-activated catalase-encoding katE gene (9). This phenotype provides a cogent explanation for the observation that CAMPs promote expression of RpoS-dependent genes and RpoS-dependent resistance to H2O2 (65), which was reported before the discovery of iraP as a PhoP-activated gene specifying a product that protects RpoS from proteolysis.
In E. coli, the iraP gene product also stabilizes RpoS (245) but is not under transcriptional control of PhoP (9). E. coli harbors a different antiadaptor gene—designated iraM—that is activated by PhoP and enables RpoS protection under the Mg2+ starvation conditions that activate PhoQ (98). There is no iraM gene in Salmonella. Thus, E. coli and S. Typhimurium utilize different PhoP-activated gene products to stabilize RpoS under similar inducing conditions. The S. Typhimurium iraP (9) and E. coli iraM (59) share the property of being induced when bacteria experience highly activating conditions for PhoQ (i.e., very low Mg2+).
The PhoP-activated mgtCBRU-cigR operon specifies products that have opposite effects on the stability of PhoP-activated proteins. On the one hand, mgtR specifies a small adaptor protein required for degradation of the inner membrane proteins MgtA (246), MgtB (169), and MgtC (154) by the inner membrane protease FtsH (247). On the other hand, mgtU specifies a peptide that protects MgtB, but neither MgtA nor MgtC, from FtsH (169). The mgtCBRU-cigR operon provides a singular example of a transcription unit specifying protease substrates, as well as proteins that further or hinder degradation of those substrates. The mgtU-dependent protection of the MgtB protein is required for S. Typhimurium both to survive inside macrophages harboring a functional Slc11a1 gene and to achieve wild-type growth after extended incubation in low-Mg2+ media (169).
Two-dimensional gel electrophoresis followed by spot identification by mass spectrometry identified a role for PhoP/PhoQ in the generation of truncated forms of DnaK, EF-G, EF-Tu, and ribosomal protein S1 (248). Curiously, these truncated proteins were observed when the inducing signal was low Mg2+ or in a strain with a constitutively active PhoQ, but not when the inducing signal was mildly acidic pH, which also activates the PhoP/PhoQ system (248). This represents another example in which different PhoQ-activating signals elicit different bacterial responses. (PhoP control of proteases other than the five discussed above is presented below under “PhoP-Dependent Modifications of the Bacterial Cell Envelope.”)
THE BEHAVIORS THAT PhoP/PhoQ REGULATES: VIRULENCE, RESISTANCE TO ANTIMICROBIAL AGENTS, MOTILITY, AND ABILITY TO COPE WITH NUTRITIONAL STRESS
In this section, we discuss the phenotypic consequences resulting from PhoP-regulated changes in gene products and metabolites. The PhoP/PhoQ system plays a critical role in the ability of several pathogens to cause disease in a variety of animal and plant hosts (17, 27, 31, 32, 249,–253). PhoP/PhoQ alters the chemical properties of the LPS (36, 44), thereby increasing resistance to a variety of CAMPs (36, 43, 139), and changes the form of motility in S. Typhimurium (254, 255) (Fig. 1). Critically, different PhoQ-activating signals often give rise to different phenotypic outcomes despite altering expression of the same PhoP-regulated genes (203, 256).
PhoP Promotes Expression of Both Virulence and Antivirulence Genes
Inactivation of the phoP or phoQ genes severely attenuates virulence in several pathogens of animals and plants (17, 27, 31, 32, 249,–253, 257). In the case of S. Typhimurium, the PhoP/PhoQ system is necessary for virulence in a variety of warm-blooded mammalian hosts (258,–260). Acquired following the consumption of contaminated water or food, S. Typhimurium experiences a strongly acidic pH in the stomach before progressing to the small intestine of a mammalian host. At the initial stages of infection, S. Typhimurium adheres to and penetrates intestinal epithelial cells. After traversing the epithelial cell barrier, S. Typhimurium is phagocytosed by macrophages or dendritic cells (DCs), which are thought to be the major determinants of systemic infection because they enable the bacterium to spread to deeper tissues via the lymphatic system and bloodstream. PhoP controls expression of virulence genes responsible for entry into epithelial cells (205), survival and replication inside phagocytic cells (17, 32, 164), and manipulation of host cell processes (261, 262). PhoP is the master regulator of S. Typhimurium virulence. Here, we discuss how the PhoP regulation of virulence and anti-virulence genes impacts the natural history of S. Typhimurium infection.
PhoP controls bacterial entry into epithelial cells.
To get into deep tissues, S. Typhimurium must first cross the gut epithelial barrier, which requires a set of bacterial proteins that elicit a host cell response resulting in bacterial internalization, a process referred to as invasion. Delivery of these proteins into epithelial cells is carried out by the type III secretion system encoded in SPI-1 (263, 264). PhoP decreases the amounts of several proteins encoded in SPI-1 (204, 205) directly, by repressing transcription of the hilA gene (206), which specifies the major transcriptional activator of SPI-1 genes (265), and indirectly, by reducing the amounts of the HilA activators HilD and RtsA (206) and by promoting transcription of the sRNA PinT that hinders translation of the hilA and rtsA genes (207). This is the reason why the hyperactive pho-24 allele renders S. Typhimurium defective for invasion of epithelial cells (205) and why a phoP-null mutant enters epithelial cells better than the wild-type strain (266).
PhoP also decreases epithelial cell invasion by repressing transcription of flagellum-encoding genes, thereby preventing flagellum-mediated motility (254, 267), which aids bacterial invasion (268). Mutants defective in SPI-1 or flagellar genes are attenuated for virulence in mice when inoculated orally (205, 268). However, it is presently unknown whether PhoP repression of invasion and flagellar genes is necessary for virulence.
Surprisingly, PhoP is also a direct transcriptional activator of genes implicated in invasion of epithelial cells. First, PhoP binds to a DNA region located within the orgA coding region in SPI-1, promoting transcription of the downstream orgB and orgC genes (269). The orgB and orgC genes (270) are the last two genes of a PhoP-repressed polycistronic transcript that includes orgA and four additional preceding genes (269). The orgB and orgC genes specify a component of the type III secretion machine (271) necessary for invasion (269) and a protein that aids assembly of the machine (272), respectively. Unlike the other five genes in the polycistronic mRNA, orgB and orgC are also transcriptionally induced under conditions that stimulate expression of SPI-2 genes (273) and inside macrophages (274), raising the possibility of orgB and orgC contributing to S. Typhimurium survival inside mammalians cells. The PhoP binding site required for transcriptional activation of the orgB and orgC genes in S. Typhimurium is not conserved in S. bongori (269), a Salmonella species that harbors SPI-1 but lacks SPI-2 (275) and is unable to survive inside macrophages (276). Second, PhoP activates transcription of grhD1, a gene that promotes bacterial invasion into mammalian cells as well as intestinal inflammation (197). PhoP activation of grhD1 is independent of that exerted by the SPI-1-encoded activator HilD (197).
In sum, PhoP represses transcription of most genes required for invasion. However, that PhoP activates some invasion genes raises the possibility of the latter genes participating in additional functions. Alternatively, or in addition, the antithetical PhoP regulation of different SPI-1 genes may allow S. Typhimurium to adapt to the continuously changing gut environment. For instance, the mammalian intestine harbors signals exerting opposite effects on PhoQ: long-chain unsaturated fatty acids decrease PhoP activity (106, 109), whereas hyperosmolarity promotes it (96), albeit transiently.
PhoP controls bacterial survival and replication inside macrophages.
A critical feature of the systemic infection caused by S. Typhimurium in mice is the ability to survive and replicate in the hostile environment of macrophages (164). PhoP plays a crucial role in this process because inactivation of the phoP or phoQ genes impairs S. Typhimurium survival and replication inside macrophages (17, 32, 164). The vast majority of S. Typhimurium resides within a phagosome—referred to as a Salmonella-containing vacuole (SCV)—when inside macrophages (277, 278), an environment that activates PhoP/PhoQ (83). Below, we discuss the different PhoP-regulated genes that contribute to S. Typhimurium survival or replication inside macrophages.
The PhoP-activated gene mgtC specifies an inner membrane protein necessary for intramacrophage survival and virulence in mice. An mgtC mutant is attenuated but less so than a phoP-null mutant (17, 27, 31, 32, 158, 164), in agreement with the notion that PhoP controls expression of other intramacrophage survival genes. Because MgtC interacts with several bacterial proteins, it is presently unclear which of these interactions are critical for virulence. First, PhoP inhibits fusion of the SCV with lysosomes, thereby hindering the delivery of various bactericidal compounds (279, 280). Although this process is mediated, in part, by MgtC (281), it is presently unclear how MgtC hampers SCV trafficking and fusion with lysosomes. Second, MgtC inhibits Salmonella’s own F1F0 ATP synthase, thereby reducing the bacterial ATP concentration and hindering acidification of the bacterial cytoplasm when S. Typhimurium experiences a mildly acidic phagosome (150). This inhibition is critical for intramacrophage survival because mgtC mutants defective in inhibiting the ATP synthase (150) are also defective in intramacrophage survival (282). A reduction in ATP concentration impacts a variety of ATP-consuming processes, including protein synthesis (38), cellulose biosynthesis (283), proteolysis by ATP-dependent proteases (37, 239), and protein solubility (284). In addition, it slows down growth, resulting in increased resistance to antimicrobial agents (285,–287).
Third, MgtC is necessary for bacterial growth in low Mg2+ conditions (158) and survival inside Scl11a1+/+ macrophages (288). Fourth, MgtC protection of the PhoP protein from proteolysis by ClpSAP is necessary for transcription of a subset of PhoP-activated genes (58), including some affecting virulence. And fifth, MgtC reduces expression of bcsA, an antivirulence gene that specifies the cyclic-di-GMP-dependent cellulose synthase (283). Although cellulose is a main component of bacterial biofilms (289), wild-type S. Typhimurium produces cellulose when inside a macrophage phagosome (283), which reduces virulence by unknown means. (Paradoxically, it has been proposed that MgtC limits S. Typhimurium pathogenicity by increasing phosphate uptake [290]. However, the latter observation is in conflict with that of others demonstrating that MgtC actually inhibits phosphate uptake [151]. The mechanisms by which PhoP controls phosphate uptake are discussed below under “The PhoP-Dependent Connection among Low Cytoplasmic Mg2+, Protein Synthesis, and the Phosphate-Responding PhoB/PhoR System.”)
The MgtC-dependent functions are designed to take place over a discrete time window because three PhoP-regulated genes limit MgtC amounts or activity. First, the antisense RNA AmgR decreases the stability of the mgtC mRNA (176), the peptide MgtR promotes degradation of the MgtC protein (154), and the protein CigR binds to MgtC, preventing MgtC from decreasing ATP amounts and from stabilizing the PhoP protein (153). Unlike the strict dependence that the mgtR and amgR genes have on PhoP for transcription, the cigR gene is transcribed from both the PhoP-activated mgtC promoter and a PhoP-independent promoter (153), which provides a basal level of CigR protein that MgtC must surpass to perform its function.
The amgR and cigR genes are anti-virulence genes because their inactivation increases S. Typhimurium virulence (153, 160, 176). By contrast, an mgtR deletion strain is minimally defective in both intramacrophage survival and virulence in mice (154, 176); this is in spite of the fact that MgtR has a much larger impact on reducing MgtC abundance than AmgR does in bacteria grown in low-Mg2+ laboratory media (176). (Nonphysiological mgtR overexpression reduces S. Typhimurium growth inside macrophages [154], which may be due to effects on both MgtC [154] and MgtB [169].)
The MgtC N92T variant is defective in interacting with CigR (153). However, a strain expressing the MgtC N92T variant instead of the wild-type MgtC protein behaves like the mgtC-null mutant rather than the cigR-null mutant inside macrophages (150, 153, 240, 282). The behavior of the MgtC N92T-expressing strain may reflect that this MgtC variant is also defective in binding to and inhibiting the F1F0 ATP synthase (150). (MgtC has a higher affinity for the F1F0 ATP synthase than for the CigR protein [153].) Though this raises the possibility of a reduction in ATP concentrations being the major way that MgtC promotes S. Typhimurium virulence, an alternative possibility is that the N92T substitution hampers other functions, such as protection of PhoP from degradation by ClpSAP (58) or interaction with the virulence protein MgtB (150).
MgtC is necessary for virulence in several pathogens, such as Mycobacterium tuberculosis, Yersinia pestis, Burkholderia cenocepacia, and Brucella suis (291,–294), that remain within a mildly acidic phagosome while inside macrophages, like S. Typhimurium. Because some of these species lack PhoP, MgtB, and/or CigR, but they all have F1F0 ATP synthase, MgtC control of the latter may be responsible for virulence in these species.
The PhoP-activated ugtL is a Salmonella-specific gene that specifies an inner membrane protein required for normal PhoP activation by mildly acidic pH, such as inside a macrophage phagosome, and for virulence in mice (87). As discussed above, full PhoP activation in mildly acidic pH requires UgtL binding to PhoQ and stimulating PhoQ autophosphorylation from ATP (87). In addition, a ugtL-null mutant is hypersusceptible to the CAMP magainin-2 and defective in modifying the LPS in ways similar to those of a phoP mutant when grown in low Mg2+ (295). Thus, UgtL appears to confer distinct functions depending on the specific signal activating PhoQ.
PhoP promotes transcription of SPI-2 genes. Although different groups reported various degrees of PhoP contribution to SPI-2 gene expression in vitro (77, 88, 161, 296, 297), the disparate results seem due to both the utilization of different reporters and bacterial growth conditions. PhoP directly activates transcription of the ssrB gene (77) (Fig. 4), thereby promoting an increase in ssrB mRNA levels that is necessary for expression of SPI-2 genes inside macrophages (77, 88, 200). PhoP also enhances SpiR abundance in acidic pH in vitro (298). In addition, PhoP directly promotes transcription of the slrP and steA genes, specifying effector proteins translocated into the host cytoplasm (102, 299) and required for virulence (300, 301).
A mutation of the PmrA-activated gene pbgP2 (also called pmrF), involved in modification of the lipid A phosphates with L-Ara4N (139), attenuates S. Typhimurium virulence in BALB/c mice inoculated orally but not inoculated intraperitoneally (302), suggesting a role for pbgP2 in survival in the gastrointestinal tract. Curiously, a pmrA-null mutant is less attenuated than the pbgP2 mutant following oral inoculation of BALB/c mice (302). By contrast, a pmrA-null mutant is hypervirulent in C3H/HeN mice inoculated intraperitoneally (215). The latter results argue that PmrA reduces expression of virulence genes and/or increases expression of antivirulence genes.
The PhoP-activated mig-14 gene mediates resistance to the CAMPs protegrin-1 and CRAMP (21, 74) and virulence (74, 303). mig-14 inactivation reduces survival and replication inside macrophages ~40% when macrophages are activated with gamma interferon and LPS (74). By contrast, the phoP-null mutant is defective for intramacrophage survival even in macrophages not activated by these agents (32, 164). The mig-14 mutant exhibits a colonization defect in mouse spleen >5 days after infection (74), suggesting that Mig-14 functions at later stages of infection.
The PhoP-activated horizontally acquired pcgL gene specifies a periplasmic d-Ala-d-Ala dipeptidase (304) and is highly induced inside macrophages (274). An S. Typhimurium pcgL-null mutant is hypervirulent in mice, a phenotype detectable at later times after infection (i.e., >7 days following intraperitoneal inoculation) (305). The hypervirulence phenotype is due to accumulation and release into host tissues of the PcgL substrate and peptidoglycan precursor d-Ala-d-Ala. Thus, the pcgL-null mutant renders wild-type S. Typhimurium hypervirulent when mice are coinfected with both strains. Moreover, hypervirulence is overcome when the pcgL mutant expresses a d-Ala-d-Ala importer from E. coli, which hinders d-Ala-d-Ala release into host tissues by importing this dipeptide into the S. Typhimurium cytoplasm (305). In addition, injection of synthetic d-Ala-d-Ala increased the virulence of wild-type S. Typhimurium in mice (305). By reducing d-Ala-d-Ala, PcgL enables S. Typhimurium to slow down replication during late stages of infection. Many natural isolates of S. enterica harbor pcgL-null mutations (305), which may give rise to increased pathogenic behaviors.
In contrast to the antivirulence proteins discussed above, which decrease the abundance or availability of virulence factors, PcgL controls the abundance of a metabolite—d-Ala-d-Ala—that acts on the host immune system. The pcgL mutant provides a singular example of how the outcome of a competition experiment can be dramatically impacted by a product accumulated by a mutant strain.
The PhoP-activated genes pagC, pagD, and pgtE are highly induced inside macrophages (274, 296) and implicated in resistance to CAMPs and serum, as well as in virulence (32, 67, 74, 303, 306,–309). However, the virulence attenuation of pagC and pagD mutants is only observed in strains harboring transposon insertions in these genes. These transposon insertions appear to confer a dominant-negative phenotype because the pagC mutant could not be complemented by a plasmid expressing a wild-type copy of the pagC gene (306) and also because a strain with the pagD gene deleted retained wild-type virulence (307). Also, lack of PgtE does not alter Salmonella survival and replication inside macrophages (310, 311), although it reduces colonization in mouse organs (312).
The Salmonella-specific PagC and PgtE confer protection from killing by serum (67, 308, 309, 313). However, the phagosomal compartment is hardly accessible to serum, raising questions as to the role that serum resistance plays in bacterial survival inside macrophages. The gut is a more likely environment for an encounter with complement, which attaches to the bacterial surface and results in efficient phagocytosis. Alternatively, or in addition, PgtE may promote bacterial dissemination by enhancing plasmin and metalloproteinases (312), aiding migration of mammalian cells (314). PgtE likely functions after S. Typhimurium is released upon macrophage death, given that its sequelog CroP has optimal activity at pH 7 (315). Critically, a single nucleotide substitution in the pgtE promoter that results in higher PhoP-independent pgtE expression results in hypervirulence in S. Typhimurium (309).
The Mg2+ transporter MgtB is necessary for growth in very low Mg2+ and for virulence in Scl11a+/+ macrophages (169) and mice (316) but is dispensable in Scl11a1–/– macrophages (169). Unlike the more ubiquitous Mg2+ transporter MgtA, which is active at both 20°C and 37°C, MgtB is only active at 37°C, reinforcing the notion that it plays a role solely in warm-blooded hosts (35).
The PhoP-dependent ability to resist CAMPs appears to contribute to S. Typhimurium’s intramacrophage survival and growth because the phoP mutant is less defective inside macrophages deficient in the antimicrobial peptide CRAMP than in CRAMP-proficient macrophages (73). PhoP’s role in CAMP resistance is largely mediated by cell envelope modifications discussed below under “PhoP-Dependent Modifications of the Bacterial Cell Envelope.”
PhoP controls macrophage cell death.
Salmonella induces programmed macrophage cell death (317,–319), a process that appears to be part of the host immune response given that mice deficient in this process (e.g., caspase-1-deficient) are hypersusceptible to Salmonella infection (320). PhoP controls macrophage cell death because the phoP mutant displays reduced macrophage death (261). This process displays distinct consequences, depending on the conditions used to grow the bacterium before contacting the macrophages (261, 317,–319, 321, 322). When cultured in LB broth to logarithmic phase, PhoP-repressed genes are responsible for early cell death (261, 318, 319, 321). When grown in the same media but to stationary phase, PhoP-activated genes are necessary for late macrophage death (317, 319), which is dependent on the PhoP-activated ssrB gene (77, 322). Although PhoP promotes macrophage death via SsrB, PhoP also reduces cell death by repressing ssrB transcription via PmrA (40, 215). The two antagonistic effects exerted by PhoP result in delayed macrophage death. Thus, PhoP controls bacterial dissemination because Salmonella-induced macrophage death appears to be responsible for bacterial dissemination in the mammalian host (323, 324). In agreement with this notion, preventing PmrA-dependent repression of the ssrB gene accelerates cell death and results in a 2-log-fold higher colonization in the organs of Slc11a1+/+ mice (215).
PhoP activities in dendritic cells and fibroblasts.
In addition to macrophages, dendritic cells (DCs) are responsible for disseminating Salmonella via the lymphatic system (325). Salmonella does not grow in DCs but survives within them (262, 326). Salmonella survival within DCs does not appear to require PhoP or PhoP-activated SPI-2 genes (262, 326,–328), which are critical for intramacrophage survival and growth. (Different groups reported slightly varying survival phenotypes in DCs but in a minor range: some reported that strains deleted for phoP or SPI-2 genes survive like the wild type [262, 326], and others reported that the mutants are slightly defective [327, 328].) However, by promoting expression of SPI-2 genes, PhoP controls SCV maturation and trafficking in DCs (262, 326, 327), which is critical for antigen presentation by DCs (262, 328). This avoidance of DCs’ antigen presentation is not achieved by the largely nonpathogenic S. bongori (329), which lacks SPI-2 (275), supporting the notion that PhoP and its activated SPI-2 genes hinder this process by DCs. Thus, PhoP allows S. Typhimurium to disseminate to various organs while staying in DCs without stimulating the adaptive immune system.
Fibroblasts are nonphagocytic cells that limit S. Typhimurium growth (266, 330). Interestingly, a phoP-null mutant replicates more than 10-fold in fibroblasts, whereas the wild-type strain does not (266, 330). The PhoP-activated ecgA gene specifying a peptidoglycan d,l-endopeptidase that cleaves γ-d-glutamyl-meso-diaminopimelic acid is induced within fibroblasts (331). The ecgA mutant survives and replicates slightly better (i.e., <2-fold) than the wild-type strain in fibroblasts. However, the ecgA mutant is defective ~5- to 6-fold in colonizing the spleen and liver after intraperitoneal inoculation (331), suggesting that EcgA-mediated peptidoglycan metabolism contributes to intramacrophage growth. Despite the opposite phenotypes of the phoP mutant in macrophages and fibroblasts (17, 32, 164, 266, 330), PhoP also promotes expression of virulence factors induced inside macrophages within fibroblasts in an acidic pH-dependent manner (330).
PhoP-Dependent Modifications of the Bacterial Cell Envelope
The composition of the cell envelope varies across bacterial species and in response to environmental cues in a given species (332). The inner membrane and the inner leaflet of the outer membrane are composed primarily of phospholipids. By contrast, the outer leaflet of the outer membrane is occupied mostly by LPS (333). The LPS is typically subdivided into lipid A, core oligosaccharide, and O-antigen (333), all of which can be modified in response to environmental conditions to serve various purposes (333). Both the inner and outer membranes have proteins embedded in them.
Maintenance of the envelope is crucial for bacterial survival because phospholipids ensure membrane integrity, the peptidoglycan is immunogenic, and the LPS is a key molecule in bacterium-host interactions, since it is at once a virulence factor, an immunogenic molecule, and a barrier to a variety of antimicrobial agents. PhoP controls the composition of the cell envelope in various Gram-negative species, in many cases via PmrA, which constitutes the major regulator of LPS modifications in S. Typhimurium (127). Below, we discuss the PhoP-dependent modifications in LPS structure and phospholipid composition, as well as the physiological roles of these modifications (Fig. 7).
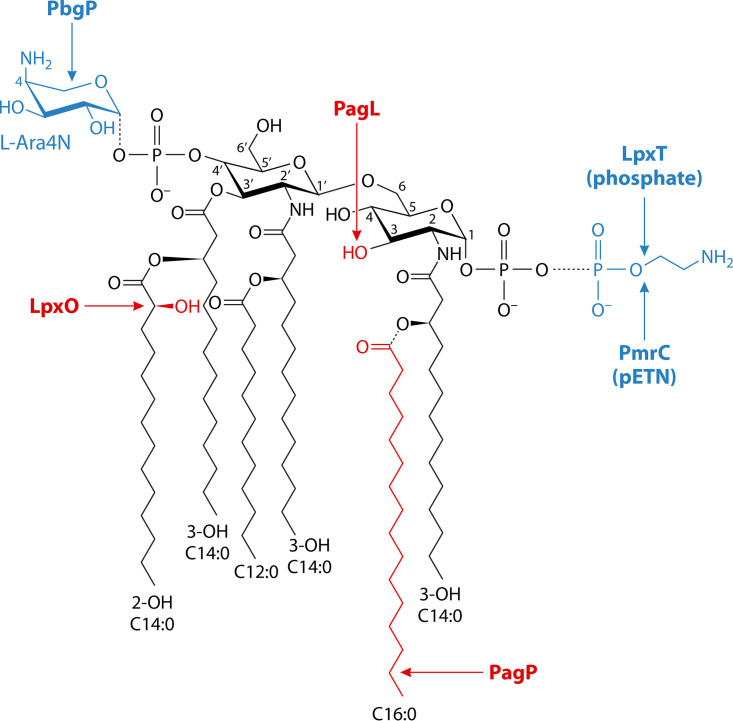
PhoP-regulated lipid A modifications in S. Typhimurium. LpxT incorporates a phosphate at position 1 of lipid A, making it a diphosphate. The LpxT-dependent phosphorylation of lipid A hinders the PmrC-dependent incorporation of phosphoethanolamine. The PbgP (Pmr, Arn) proteins incorporate 4-amino arabinose to position 4′ of lipid A (as depicted) and/or position 1. PagL removes an R-3-hydroxymyristate group from lipid A. LpxO hydroxylates lipid A. PagP incorporates a palmitoyl group to lipid A.
The PhoP-activated pagL gene from S. Typhimurium (10) specifies an outer membrane protein that removes the R-3-hydroxymyristate group from lipid A (334) (Fig. 7). A PagL sequelog is found in P. aeruginosa but not in E. coli or Y. pestis (335). The PagL-dependent deacylation is detectable only when the PmrA, Ugd, or PbgP2 protein is absent (336), indicating that the L-Ara4N modification of lipid A prevents deacylation. PagL’s substrate specificity is mediated by arginine residues at positions 43, 44, and 135 (337). A strain expressing the PagL R43A variant displayed increased permeability (338), suggesting that PagL-mediated deacylation enhances resistance to lipophilic antimicrobial compounds (338); by contrast, PagL overexpression has no effect on antibiotic resistance. The PagL-catalyzed lipid A modifications reduce the Toll-like receptor 4 (TLR4)-dependent immune response (339), indicating that PagL plays a role in immune evasion. These results suggest that the PagL-dependent modification takes place under conditions that activate the PhoP protein but fail to activate the PmrA protein.
The inner membrane protein LpxO is responsible for the lipid A hydroxylation, resulting in lipid A modified with 2-hydroxymyristate (340, 341) (Fig. 7). Flanked by two transmembrane domains, the LpxO enzymatic region faces the cytoplasm. Lipid A hydroxylation and lpxO transcription are PhoP dependent. An LpxO sequelog is found in P. aeruginosa but not in E. coli or Y. pestis (335).
A large number of genes involved in LPS modification are controlled by PhoP indirectly, by the PhoP-PmrD-PmrA pathway in S. enterica (6, 40, 127) (Fig. 5), as discussed above under “Posttranslational Control of DNA-Binding Regulatory Proteins and Unusual PhoP Role at Certain Promoters.” These genes include ugd and those in the pbgP operon (ugd and pbgP are also referred to as pmrE and pmrH, respectively), which are responsible for the covalent modification of the lipid A phosphates with L-Ara4N (Fig. 7), and pmrC (also referred to as eptA) responsible for the covalent modification of the lipid A phosphates with phosphoethanolamine (127) (Fig. 7). PmrA-regulated genes also encode products mediating the modification of the core oligosaccharide (pmrG and cptA) and the O-antigen (wzzfepE and wzzST) (127). In some species, PhoP is a direct transcriptional activator of some of these lipid A-modifying genes that are directly regulated by PmrA in S. Typhimurium. For example, PhoP is a direct transcriptional activator of the pbgP operon and ugd gene in both Y. pestis (5) and K. pneumoniae (236).
LpxT is an inner membrane protein that takes the terminal phosphate from undecaprenyl-pyrophosphate (C55-PP) and puts it at position 1 of lipid A (342) (Fig. 7). PhoP is a direct transcriptional activator of the lpxT gene (256). That PhoP promotes lpxT expression can seem paradoxical given that PhoP mediates resistance to multiple CAMPs and that LpxT increases the overall negative charge of the LPS. Moreover, PhoP activates PmrA, resulting in expression of PmrR, a short membrane protein that binds LpxT and inhibits its lipid A-modifying activity (343). That is, PhoP promotes transcription of both the lpxT gene and of an inhibitor of the lpxT gene product.
The PhoP activation of lpxT transcription enables S. Typhimurium to differentially modify the lipid A phosphates solely with L-Ara4N or with a combination of L-Ara4N and phosphoethanolamine (Fig. 7), depending on the identity of the signal activating the sensor PhoQ: low periplasmic Mg2+ or a mildly acidic pH, respectively (256). LpxT-modified lipid A favors incorporation of L-Ara4N. This is because PhoQ activation in low Mg2+ provides lpxT with an expression kinetic advantage over pmrC (256), which is responsible for the modification of lipid A with phosphoethanolamine (232). The expression kinetic advantage reflects that the lpxT gene is directly activated by PhoP, whereas pmrC is activated by PmrA and requires PhoP to activate pmrD transcription, PmrD protection of PmrA-P, and PmrA-P binding to the pmrC promoter (40, 41). When the inducing condition is mildly acidic pH, both PhoQ and PmrB are activated, which results in similar transcription timing for lpxT, pmrC, and pmrR, and favors lipid A modification with phosphoethanolamine, as well as with L-Ara4N (256). In other words, the type of modification experienced by the lipid A phosphates is determined by the order in which lipid A-modifying genes are expressed rather than simply the expression of these genes.
When harvested from inside macrophages, the S. Typhimurium lipid A contains both L-Ara4N and phosphoethanolamine (344), supporting the notion that mildly acidic pH is the PhoQ-activating signal inside macrophages (85, 87, 203). In addition, bacteria in which the lipid A is modified with both L-Ara4N and phosphoethanolamine exhibit more resistance to polymyxin B than those modified solely with either L-Ara4N or phosphoethanolamine (256).
What is the role(s) of the PhoP-regulated LPS modifications? First, as discussed above, some of these modifications are proposed to help the bacterium evade the host immune system. Second, some modifications lower the overall negative charge of the membrane, which decreases binding of and toxicity by Fe3+ (345), as well as Fe3+-mediated activation of the sensor PmrB (343). Third, modification of the lipid A phosphates with L-Ara-4N and phosphoethanolamine confers resistance to polymyxin B (36, 139, 232). This phenotype is not simply due to making the surface less negatively charged because bacteria modified with both L-Ara-4N and phosphoethanolamine are more resistant to polymyxin B than those modified solely with L-Ara-4N even though modification with L-Ara-4N decreases the surface negative charge more than with phosphoethanolamine (256). And fourth, the lipid A modifications of phosphate residues release Mg2+ ions neutralizing those phosphates. The latter property effectively turns the outer membrane into a reversible Mg2+ storage site that can be tapped for importation into the cytoplasm by the PhoP-activated transporters MgtA and MgtB (36).
Beyond regulating LPS-modifying proteins, PhoP controls phospholipid composition and structure. The inner membrane protein PbgA (known as YejM in E. coli) is particularly interesting in this regard because it functionally connects LPS and phospholipids. PbgA binds to the outer membrane in a PhoP/PhoQ-dependent manner (346). An increase in cardiolipins in the outer membrane is dependent on PbgA’s periplasmic domain (346). Two arginine residues at positions 215 and 216 of PbgA are required for outer membrane association and bacterial growth (346). Lack of the periplasmic domain of PbgA increases antibiotic susceptibility and reduces survival inside macrophages (346) (note that deletion of PbgA’s transmembrane domains is lethal). Furthermore, the strain lacking PbgA’s periplasmic domain harbors increased amounts of lipid A-core molecules, cardiolipin, and phosphatidylethanolamine and decreased amounts of specific phospholipids with cyclopropanated fatty acids (347). Inactivation of the lapB and lpxC genes, involved in LPS biosynthesis, restores pbgA mutant phenotypes, such as phospholipid and LPS assembly defects (347). The lack of LpxC also renders the pbgA mutant’s virulence closer to that of wild-type S. Typhimurium in TLR4-deficient mice (347), which are defective in recognition of lipid A. The latter findings are further supported by the crystal structure of the LPS-PbgA complex (348). By binding to lipid A, PbgA controls LpxC degradation by the protease FtsH, thereby modulating LPS metabolism (348). This is because LpxC governs the first committed step in lipid A biosynthesis. Although PbgA was proposed to serve as a cardiolipin transporter (346), the lipid A-binding portion of PgtA does not interact with cardiolipin (348), arguing against a role for PbgA in cardiolipin transport.
PhoP further controls membrane phospholipids by directly promoting transcription of the pagP gene (10), which specifies an outer membrane protein that acylates glycerophospholipids by adding palmitoyl groups (349) and then transfers these groups to lipid A (43, 350). These modifications enhance resistance to C18G (43), magainin 2, and cecropin A (295), but not to defensin NP-1 (351) and polymyxin B (295). Coincidentally, the first three CAMPs are all alpha-helical peptides, while defensin NP-1 is a beta-sheet peptide, and polymyxin B is cyclical, suggesting that palmitoyl-lipid A specifically enhances resistance to alpha-helical peptides. Testing of additional CAMPs would be required to establish this as a general rule.
By contrast, E. coli is also able to perform the PagP-dependent acylation reaction when treated with metavanadate, but it does not do so under typical growth conditions (352). PagP-related proteins are present in a variety of Gram-negative species, including Legionella pneumophila (353), Bordetella bronchiseptica (354), and Y. enterocolitica (355), where they modify lipid A, increase resistance to CAMPs, and promote virulence.
As discussed above, the PhoP-activated EcgA is a peptidoglycan d,l-endopeptidase that modifies the peptidoglycan (331). PhoP promotes the expression of this enzyme when inside host cells, such as fibroblasts and epithelial cells. EcgA promotes S. Typhimurium virulence, and yet its physiological function responsible for the virulence phenotype is unclear.
PhoP-Dependent Switch from Flagellum-Mediated to Flagellum-Independent Forms of Motility
Bacterial motility allows the exploration of new niches (356). Flagella are a complex assembly that includes a motor-stator complex that powers the rotation of a filament (357), thereby allowing the bacteria to swim. Flagella are necessary for S. Typhimurium invasion into host epithelial cells (358), though they do not alter virulence (359). However, the flagellum is strongly immunogenic, being recognized by Toll-like receptor 5 (TLR5) (360) and NLRC4 inflammasome (361, 362) in mammals. Therefore, decreasing flagellum expression would lower the chances of bacterial recognition by the host immune system.
When experiencing a mildly acidic pH, S. Typhimurium decreases the amounts of the two flagellin structural proteins—FliC and FljB—in a PhoQ-dependent manner (254). This decrease reduces flagellum-mediated motility at low pH (254). Transcription of the fliC gene is reduced in the pho-24 strain (254), arguing for a transcriptional effect. By contrast, transcriptional control of the fliF and flgB genes, which specify components of the rotor and basal body, respectively, is not PhoQ dependent (254). The differential regulation of flagellar components may constitute a strategy to keep most of the flagellar machinery ready inside an acidified phagosome without displaying the immunogenic filament, avoiding a host immune response. Alternatively, or in addition, it may stem from a different requirement.
As discussed above, the sRNA originating from the leader region of the mgtCBRU-cigR operon promotes degradation of the fljB/fljA transcript (163), thereby hindering motility by flagella made with the FljB filament. A fljB mutation that renders the corresponding mRNA refractory to repression by the sRNA increases S. Typhimurium’s ability to replicate inside macrophages and increases virulence in mice inoculated intraperitoneally with a dose 1,000 ×
× the LD50 (163). Given that mutants defective in either one of the two flagellin forms behave like the wild-type strain inside macrophages (163), the advantage of repressing only one of the flagellin types is currently mysterious.
the LD50 (163). Given that mutants defective in either one of the two flagellin forms behave like the wild-type strain inside macrophages (163), the advantage of repressing only one of the flagellin types is currently mysterious.
PhoP promotes a flagellum-independent form of motility when experiencing very low Mg2+ (255). The PhoP-activated Salmonella-specific pagM gene specifies a secreted protein that mediates a flagellum- and pilus-independent motility (255). An S. Typhimurium strain that expresses PagM confers motility to a strain lacking PagM (255), suggesting that PagM promotes a group behavior. Transcriptional activation of pagM is mgtA and mgtC dependent (255), indicating that it requires large amounts of active PhoP protein and takes place in organisms in which the cytoplasmic Mg2+ concentration has decreased beyond the threshold that triggers expression of the MgtA and MgtC proteins.
The PhoP-Dependent Connection among Low Cytoplasmic Mg2+, Protein Synthesis, and the Phosphate-Responding PhoB/PhoR System
Protein synthesis is the most energy-demanding process that cells undertake, accounting for 50 to 60% of ATP consumption in bacteria (363). Protein synthesis takes place in the ribosome, a complex of multiple RNAs and proteins that is strictly dependent on Mg2+ for assembly (38, 364). When the cytoplasmic Mg2+ concentration drops below a certain threshold, protein synthesis is compromised due to inefficient ribosome assembly. PhoP governs a bacterial response to low cytoplasmic Mg2+ that enables protein synthesis to continue, albeit at a reduced rate (38). This response entails Mg2+ uptake by the PhoP-activated Mg2+ transporters MgtA and MgtB and a decrease in ATP concentration by the MgtC, MgtA, and MgtB proteins (38). The decrease in ATP concentration lowers transcription of rRNA genes, which prevents the accumulation of the negatively charged rRNA from titrating the available Mg2+ (38). This response enables protein synthesis and the assembly of some ribosomes to continue at a low rate because under most growth conditions the rate of protein synthesis is directly correlated with the number of functional ribosomes in the cell. Although E. coli lacks the mgtC and mgtB genes, it relies on PhoP to make the necessary adaptations for protein synthesis to continue under low-cytoplasmic-Mg2+ conditions (38).
The PhoB/PhoR system is activated in S. Typhimurium experiencing low cytoplasmic Mg2+. This is because low cytoplasmic Mg2+ compromises ribosome assembly, thereby decreasing the rate of protein synthesis. This lowers ATP hydrolysis and prevents the liberation of inorganic orthophosphate (Pi), which results in a decrease in free cytoplasmic Pi (38) that activates PhoB/PhoR regardless of the extracellular Pi concentration (30). This activation, in turn, promotes expression of genes mediating Pi import. This is why treatment with protein synthesis inhibitors activates the PhoB/PhoR system even in organisms harboring plenty of Pi and Mg2+ (38).
When S. Typhimurium experiences low-cytoplasmic-Mg2+ conditions, the MgtC protein decreases the cytoplasmic phosphate concentration (38) by inhibiting a yet-to-be-identified Pi importer (151). A decrease in Pi uptake is expected to aid bacteria experiencing low-cytoplasmic-Mg2+ conditions by decreasing the abundance of a major Mg2+-chelating metabolite. By contrast, the Lee group has proposed that MgtC actually increases Pi uptake by promoting PhoR autophosphorylation (290). Curiously, the latter study reported that a mutation in the catalytic domain of PhoR abolishing its interaction with MgtC had phenotypes even in noninducing conditions (290), raising the possibility of the investigated mutation disrupting the catalytic function of PhoR. Because the two studies were carried out in media containing different carbon sources (30, 290), it is possible that the different results are due to a connection between carbon and phosphate metabolisms.
In addition to the effects discussed above, MgtC reduces ATP accumulation by inhibiting the F1F0 ATP synthase (150) and synthesis of ribosome subunits, thereby balancing the cytoplasmic Mg2+ contents and ribosomal subunits to enhance translation (38). Finally, as discussed in previous sections, MgtC also promotes proteome homeostasis by reducing ATP-dependent proteolysis (37), reducing the need for synthesis of new proteins. Therefore, disruption of protein synthesis causes detrimental physiological changes that are corrected by the PhoP-activated protein MgtC, which alleviates the need for ribosomes and thus restores Pi homeostasis, ATP availability, and ribosome assembly.
PhoP-Dependent Metal Ion Homeostasis
Bacteria require a variety of metal ions as cofactors in enzymatic reactions (35, 365, 366). However, metal ions are often toxic in elevated concentrations. For instance, Fe2+ causes oxidative stress via the Fenton reaction (366). Therefore, bacteria have both import and export systems to control metal ion homeostasis (367), and some of these systems are under PhoP control.
PhoP activation in low Mg2+ promotes transcription from the promoters of both the mgtA gene and the mgtCBRU-cigR operon, which, depending on cytoplasmic signals, results in Mg2+ import via the ATP-driven MgtA and MgtB transporters (35, 142, 143). Because PhoP reverses membrane potential (368), Mg2+ cannot be imported via CorA because this protein is a channel regulated by membrane potential (35, 146).
When S. Typhimurium experiences a mildly acidic pH, PhoP promotes Fe2+ import via RstA-mediated expression of the Fe2+ importer FeoB (222). Curiously, RstA’s cognate sensor RtsB is dispensable for this activation (222), and how RstA is activated by PhoP in mildly acidic pH is not yet known. As discussed above, PhoP-P reduces EIIANtr abundance (159), which is expected to favor binding of Fur, the master regulator of iron homeostasis, to DNA (369), resulting in repression of Fur regulon genes, such as fepA, fhuA, and iroB (369), and therefore decreasing iron uptake. The existence of the full regulatory cascade remains to be demonstrated but represents a puzzling contrast with PhoP activation favoring iron uptake as observed with FeoB. This inconsistency may indicate the presence of other, currently unknown signals integrated into the regulatory networks that define whether PhoP promotes or hinders iron uptake.
This emerging connection between Mg2+ and Fe2+ is reminiscent of the connection between the Mg2+-sensing PhoP/PhoQ system and the Fe3+-sensing PmrA/PmrB system via PmrD and may reflect a physiological need to link the pools of Mg2+ and iron ions.
PhoP-Dependent Resistance to Acidic pH
Bacteria can experience acid stress during their lifestyles. In particular, enteric bacteria must cross the gastric barrier to reach the intestine, which involves resisting severe pH stress (down to pH 1.5). By contrast, bacteria typically need to maintain a cytoplasmic pH around 7 for their enzymes to function. Resistance to acid stress involves many different systems that either pump protons out of the cytoplasm or use them in certain biochemical reactions to stabilize cytoplasmic pH (for a comprehensive review, see reference 370).
In E. coli, PhoP promotes transcription of the gadE gene (11), specifying a transcriptional regulator of the E. coli-specific gadBC and gadA genes involved in acid resistance (371, 372). GadA and GadB are two cytoplasmic glutamate decarboxylases that both catalyze the conversion of glutamate and protons into GABA and CO2, thereby consuming protons (370). These proteins are coupled with the inner membrane glutamate/GABA antiporter system GadC that specifically imports neutral glutamate and exports protonated GABA, resulting in a durable removal of one proton from the cytoplasm per transport reaction (373, 374).
PhoP also promotes transcription of the E. coli-specific hdeAB operon (11), specifying periplasmic chaperone proteins (375,–377). HdeA and HdeB exist as inactive homodimers at neutral pH, but they dissociate into active monomers when cells experience an acidic periplasmic pH (375, 377). Both proteins are necessary for resistance to severe pH stresses (pH 2.0) (377) since they help in refolding proteins denatured in the periplasm due to acid stress, the accumulation of which is toxic. In addition, PhoP directly activates transcription of the hdeD gene, which is divergently transcribed from the hdeAB operon (378). HdeD is a membrane protein of unknown biochemical function that enhances acid resistance at high cell density (378). By contrast, S. Typhimurium merely harbors an hdeB-like gene, and it is not known whether its transcription is PhoP regulated.
Finally, the stress response sigma factor RpoS is critical for surviving a multitude of stresses, including sustained acidic pH (379). In E. coli, this ability is ascribed to its promotion of the transcription of the gadE, gadBC, gadA, and hdeAB genes (370). As discussed in previous sections, PhoP stabilizes RpoS via IraM in E. coli (96), thereby promoting transcription of acid resistance genes both directly and indirectly via RpoS.
Likewise, PhoP promotes acid resistance in Salmonella. When exposed to a mildly acidic pH (pH 6), S. Typhimurium becomes up to 1,000-fold more resistant to the subsequent challenge of a more acidic pH (pH 3) compared to cells that did not experience the mildly acidic pH (380). This mechanism is termed adaptive Acidification Tolerance Response (aATR). The phoP mutant is more sensitive to acidic pH challenge in both nonadapted and aATR conditions (380). The molecular mechanisms involved are less well characterized than in E. coli. In S. Typhimurium, resistance to organic acids requires both PhoP and RpoS (81, 379). Although PhoP increases RpoS amounts by stabilizing it via IraP (9), the rpoS mutant is defective in organic acid resistance even in aATR conditions where the phoP mutant displays resistance to acid stress (albeit not as much as wild type) (81). The rpoS phoP double mutant displays additive effects by RpoS and PhoP on acid resistance (81). These findings indicate that PhoP has multiple ways to render the organism resistant to acid stress, one via RpoS, and others currently unknown. The CadA/CadB system may be one such effector of PhoP-mediated acid stress resistance in S. Typhimurium. CadA is a lysine decarboxylase and CadB a lysine/cadaverine antiporter that contributes to acid resistance in both E. coli (370) and S. Typhimurium (381). In E. coli, the gene silencer H-NS represses cadBA expression (382). If a similar silencing takes place in S. Typhimurium, the PhoP-mediated H-NS degradation occurring in mildly acidic pH (203) may contribute to acid resistance via cadBA.
The relationship between S. Typhimurium and acidic pH is notable. As discussed above, the presence of an acidic environment is actually beneficial for S. Typhimurium. Acidification of the SCV is required for S. Typhimurium replication in both macrophages (82) and nonphagocytic cells (330), for PhoP/PhoQ activation inside macrophages (83, 166), and for virulence in mice (383). Curiously, the phoQ mutant grows better than the wild-type strain in mildly acidic pH (pH 5.5) (63), whereas it is defective in resistance to strongly acidic pH (pH 3) (81). This implies that a system other than PhoP/PhoQ is responsible for adaptation to mildly acidic pH and that PhoP/PhoQ is specialized in resistance to lower values. Considering that a large portion of the S. Typhimurium PhoP regulon does not mediate resistance to acidic pH, low pH represents both a cue to mount resistance mechanisms and a signal required to induce PhoP regulon members that do not participate in resistance to acidic pH.
PhoP Is Required for Virulence in Bacteria Causing Different Disease Conditions
Below, we discuss phoP’s involvement in the virulence of a wide variety of bacterial pathogens. It is striking that PhoP controls virulence in bacteria that cause different disease conditions, proliferate in different tissues and organs, and exhibit distinct host specificities. Typically, the PhoP-regulated genes responsible for virulence are pathogen specific, reflecting the impact of horizontal gene transfer on the evolution of bacterial pathogens, the need of bacterial pathogens to coordinate expression of horizontally acquired virulence genes with those in the ancestral genome, and often, mediation of this coordination by the ancestral PhoP/PhoQ system.
S. enterica is the best-understood bacterial species regarding PhoP’s involvement in virulence. As discussed above, PhoP governs the typhoid-like disease that S. Typhimurium causes in mice. In S. Choleraesuis, inactivation of the phoP gene attenuates virulence in pigs (259), the natural host of this S. enterica serovar. In the human-adapted serovar Typhi, a derivative deleted for the phoP and phoQ genes was safe and immunogenic in human volunteers (260). The phoP and phoQ genes are required for expression of the Typhi-specific genes encoding typhoid toxin (384). (S. Typhi mutants in phoP or in the typhoid toxin-specifying genes displayed a wild-type virulence in hu-SRC-SCID mice inoculated intraperitoneally with a very high bacterial dose [385], suggesting that this animal model does not reflect the behavior observed in humans, the natural host of S. Typhi.)
The phoP and phoQ genes are required for virulence in chickens in the host-restricted serovar Gallinarum (258). This is also the case for phoP in the broad host range serovars Typhimurium and Enteritidis (386). That phoP mutants of serovars Typhimurium and Enteritidis retained a wild-type ability to colonize the intestine of chickens (386) and inhibited Salmonella colonization demonstrates their potential as vaccine strains by themselves and when harboring additional mutations in other virulence genes (386, 387). The vaccine potential of a phoP mutant S. Typhimurium was first identified in one of the three papers that implicated phoP in virulence (31).
A phoP mutant S. Typhimurium is attenuated not only in its natural animal hosts but also in widely different models of infection. First, a phoP-null mutant was attenuated following intravenous infection of zebrafish (388), albeit not as attenuated as a purA mutant, which is defective in purine metabolism (388). The phoP mutant displayed increased virulence upon inhibition of an autophagy-related pathway that is dependent on the Rubicon protein from zebrafish (388). In contrast to the essential virulence role that the PhoP-activated SsrB/SpiR system plays in mice (209, 210), an ssrB S. Typhimurium mutant retained wild-type virulence in zebrafish (388). And second, both a strain from which the phoP, phoQ, and purB genes were deleted and one with the pho-24 allele produced less killing in the nematode Caenorhabditis elegans than the isogenic wild-type strain (389). Knocking down the production of antimicrobial peptides in C. elegans restored gut colonization by a phoP-null mutant (390).
Shigella flexneri is an intracellular pathogen that, like S. Typhimurium, is capable of infecting epithelial cells and macrophages. A phoP mutant S. flexneri is attenuated for virulence in both mice and guinea pigs (249, 250). Similar to what is observed in S. Typhimurium, lack of phoP or phoQ results in defective growth in low Mg2+ and reduced resistance to antimicrobial peptides (249, 250). By contrast, the secreted protein effectors responsible for entry into host cells differ between the two species (391). Encoded in its large virulence plasmid, the Shigella-specific gene icsA is PhoP activated and specifies an outer membrane protein mediating invasion of epithelial cells and virulence in guinea pigs (250). PhoP controls expression of different horizontally acquired virulence genes in S. flexneri and S. enterica, indicating that the control of foreign virulence genes by PhoP emerged several times during the evolution of enteric bacteria.
That control of foreign virulence genes by PhoP arose independently in different lineages is further supported by the transcriptional activation that PhoP exerts on lmiA, a gene specific to enterohemorrhagic E. coli (EHEC O157 H7) and necessary for virulence (257). The LmiA protein, in turn, promotes expression of Ler, the master regulator of genes in the locus of enterocyte effacement (LEE pathogenicity island) (257). The LmiA-mediated activation that PhoP exerts on LEE is independent of that exerted by MgrR, the PhoP-activated regulatory RNA (59) that enhances expression of the regulatory protein GrlA, an activator of one of the LEE operons (392).
Because the small intestine is the primary site of Mg2+ absorption in the mammalian gut, Mg2+ levels are expected to be low in the large intestine, suggesting that low Mg2+ is the activating signal for the EHEC PhoP/PhoQ system during infection (257). In agreement with this notion, feeding magnesium to mice reduced EHEC adherence to the mouse intestinal tract (257). In addition, the mildly alkaline pH of the large intestine makes EHEC PhoQ activation by mildly acidic pH improbable. Unlike EHEC activation of PhoP/PhoQ in the large intestine, a mildly acidic pH is the signal that activates the PhoP/PhoQ system when S. Typhimurium is inside macrophages (85, 87).
As discussed above, phoP is necessary for survival against acid stress in both S. enterica and E. coli. By contrast, PhoP is dispensable for acid stress survival in Shigella (249), which is surprising due to the close relatedness between Shigella species and E. coli (strictly speaking, Shigella and E. coli belong to the same species because their DNAs can readily recombine).
Inactivation of the phoP gene in Y. pestis and Y. pseudotuberculosis renders these organisms defective for intramacrophage survival and virulence in mice (251, 393). However, the observed attenuation is not as pronounced as that observed in S. Typhimurium, which may reflect the distinct behaviors of the two pathogens inside host cells. For example, unlike what takes place in S. Typhimurium, the phagosomal compartment containing Y. pestis fuses with lysosomes independently of PhoP (292).
The PhoP-activated mgtC and ugd genes are required for intramacrophage survival in Yersinia species, whereas only the former is necessary for this property in S. Typhimurium. PhoP also controls expression of the pbgP operon that, together with the ugd gene, mediates lipid A modification with L-Ara4N, rendering the bacterium resistant to certain CAMPs. However, whereas the promoters of the pbgP and ugd genes are directly activated by PhoP in Y. pestis, they are activated by PhoP indirectly in S. Typhimurium, via the PhoP-PmrD-PmrA pathway (5, 394). Therefore, Y. pestis preserves some of the functions carried out by PhoP in S. Typhimurium but implements them in a different fashion.
Photorhabdus luminescens is a symbiont of nematodes and a pathogen of insects. P. luminescens requires a functional phoP gene for virulence in the insect model Spodoptera littoralis (395). As described in S. Typhimurium, the phoP mutant P. luminescens displays increased motility (395). Unlike the pronounced growth defect in low Mg2+ of a phoP S. Typhimurium (18), however, the phoP mutant P. luminescens exhibits only a minor defect (395). The P. luminescens phoP mutant is also attenuated for virulence in Drosophila melanogaster, but, interestingly, behaves like wild-type P. luminescens in a D. melanogaster imd mutant (396), which is defective in the production of CAMPs that constitute a large component of the innate immune response in this organism. Mutants defective in the phoP gene or the PhoP-activated pbgP operon are hypersusceptible to different CAMPs (397). These genes are required for the generation of a bacterial subpopulation that exhibits CAMP resistance and septicemia in insects (397).
As in S. Typhimurium, PhoP-activated genes are expressed in response to low Mg2+ in P. luminescens, and they include the pbgP operon and the mgtE gene specifying a Mg2+ transporter (397), albeit of a protein family different from that to which MgtA and MgtB belong (35). Unlike S. Typhimurium, PhoP promotes expression of cutF, a gene implicated in copper homeostasis (397).
In the phytopathogen Dickeya dadantii, inactivation of the phoP or phoQ genes moderately attenuates virulence (252, 398). The phoQ mutant is defective in surviving moderately acidic pH conditions (pH 4.5 to 5.5) and CAMPs (252, 398). Similar to what is observed in S. Typhimurium (254, 267), motility is repressed by PhoQ (399). The repression of flagellum-mediated motility appears to be the general rule, as phoP mutants of pathogenic E. coli are hypermotile as well (368). Laboratory strains of E. coli also exhibit pH-dependent flagellar motility (400), but the involvement of PhoP remains to be tested. In contrast to the phenotype displayed by the bacterial species discussed above, a phoP mutant has reduced motility in the phytopathogenic bacterium Xanthomonas (401), reflecting that PhoP regulates a wide variety of flagellar genes, including those specifying motor proteins (401). Thus, PhoP activation of motility in Xanthomonas likely represents a species-specific adaptation rather than a general rule.
In Pectobacterium spp, the PhoP/PhoQ system (sometimes referred to as PehR/PehS) causes a variety of diseases in different plants. In P. versatile, the PhoP/PhoQ system is regulated by the divalent cations Ca2+ and Mg2+ (402) as in S. Typhimurium (18). The most strongly unregulated gene by PhoP is pehA (402), which specifies the major endo-polygalacturonase, a critical virulence factor required to degrade components of the plant cell wall. PhoP control of pehA is conserved in other Pectobacterium spp., such as P. parmentieri (403). Transcriptomic and bioinformatic analyses revealed that the vast majority of PhoP-regulated genes in P. versatile are not regulated by PhoP in other bacterial species and that the motif recognized by PhoP is largely conserved (402). This property likely reflects the distinct lifestyles and environments explored by the different PhoP-containing species. In P. versatile, for example, PhoP strongly represses transcription of genes mediating arabinose transport and utilization but increases production of an N-acyl homoserine lactone (402). These nutritional and quorum molecule-producing behaviors have not been reported in PhoP-containing species that infect animals.
Like infection of animal hosts by S. Typhimurium, the infection of plants by Pectobacterium spp. is a dynamic process in which the infecting bacterium experiences a range of host environments that often change in response to activities carried out by the infecting bacterium. PhoP-activated genes are highly expressed at the onset of infection (402), but the activation wanes at later stages of infection (404). Decreased expression of the PhoP-activated phoP gene occurs in macerated potato tubers (402) and in infected tobacco stems (405).
A phoP-null mutant of the fish pathogen Edwardsiella tarda was attenuated for virulence following intramuscular injection of zebrafish (406). The phoP mutant was also hypersensitive to several antibiotics and antimicrobial peptides and grew more slowly than the wild-type strain in the second phase of diauxic growth in complex media (406). A comparative proteomics experiment identified 31 proteins whose abundance differed between wild-type and phoP mutant E. tarda strains. AtpD, one of the proteins that is part of the ATP-generating machine F1F0 ATPase, exhibited lower abundance in the phoP mutant than in the wild-type strain (407), reflecting differences in the mRNA abundance of the corresponding gene. Activation of atpD, the eighth gene in the atpIBEFHAGDC operon, is ascribed to PhoP binding to the promoter that precedes atpI, the first gene in the operon (407). The F1F0 ATPase both makes and breaks down ATP (408), and it is presently unclear whether the PhoP-dependent upregulation of the atpIBEFHAGDC operon alters ATP amounts and, if so, in which direction. As discussed above, PhoP promotes a decrease in ATP amounts in S. Typhimurium (38, 150).
A phoQ mutant of the environmental organism and broad host range pathogen P. aeruginosa is impaired for virulence in lettuce, epithelial cells, and mice (253). Curiously, phoP and phoQ mutants display different behaviors, as some genes appear to be regulated only by PhoQ and not by PhoP (253). Although P. aeruginosa PhoQ was previously proposed to mediate only dephosphorylation of PhoP-P (125), it functions like the S. Typhimurium PhoQ when expressed in S. Typhimurium (86). The P. aeruginosa PhoQ is also necessary for host cell death, which is potentially mediated via increasing production of proteases and lipases and a less immunogenic LPS (409). The opportunistic nosocomial pathogen Stenotrophomonas maltophilia harbors a Mg2+-regulated PhoP/PhoQ system that promotes resistance to a variety of antibiotics (410). This is in addition to resistance to polymyxin B that the PhoP/PhoQ system promotes also in other human pathogens.
EVOLUTION OF PhoP AND ITS REGULON
PhoP/PhoQ: a System Specific to Proteobacteria?
The PhoP/PhoQ system is widespread across most of the commonly studied gammaproteobacterial genera with the notable exception of Vibrio. Agrobacterium spp. appear to have phoP/phoQ orthologs (NP_353994.2 shares 43% amino acid identity with the S. Typhimurium PhoP), but no experimental data are available for the PhoP/PhoQ system in this organism. Below, we consider only the gammaproteobacterial PhoP/PhoQ system, in which the orthology is clear. We discuss conservation in the signals controlling PhoP activity and the physiological functions governed by PhoP, the targets of PhoP regulation in terms of gene function, DNA sequences recognized by PhoP, and the promoter architecture of genes activated by PhoP.
Conservation of the Signals Controlling PhoP and PhoQ
The response to Mg2+ is conserved among all investigated PhoQ-containing bacterial species with the notable exception of the insect symbiont Sodalis glossinidus (35). In agreement with this notion, PhoP is dispensable for growth in low Mg2+ in S. glossinidus (411), which is in contrast to the PhoP requirement displayed by organisms with a Mg2+-responsive PhoQ protein. S. glossinidus’s behavior is hypothesized to result from an adaptation to the Mg2+-rich hemolymph in which this organism resides (411). This is further supported by the fact that the PhoP/PhoQ system is regulated by Mg2+ in S. praecaptivus (412), a related free-living Sodalius species.
The S. glossinidius PhoQ is activated neither by the investigated CAMPs, which may reflect that S. glossinidius exhibits constitutive resistance to CAMPs, nor by mildly acidic pH (411). Likewise, the PhoQ protein from P. aeruginosa does not respond to the synthetic mammalian-like CAMP C18G (86), and it remains unknown whether PhoQ is activated by CAMPs more likely to be encountered by P. aeruginosa in its natural habitat as an environmental organism. In the plant-associated bacterium Dickeya spp., PhoQ is activated by the plant AMP thionin (252).
Conservation of the Physiological Functions Controlled by PhoP
There is remarkable conservation in the cellular functions controlled by PhoP even when the form of regulation (i.e., direct versus indirect) or the genes mediating such functions differ across species. The observed conservation may reflect a bias for studying PhoP/PhoQ mostly in enterobacteria. PhoP control of Mg2+ homeostasis is almost universally conserved (35). This means that PhoP is a transcriptional activator of a Mg2+ transporter gene, which may not be the same gene across bacterial species. For example, PhoP is a direct transcriptional activator of the mgtA and mgtB genes in S. Typhimurium (10, 123). These two genes specify Mg2+ transporters of the same protein family (35). PhoP is also a direct transcriptional activator of the mgtA gene in E. coli (180), which lacks mgtB. In Y. pestis, which lacks mgtA, PhoP is a direct transcriptional activator of mgtB (46). In the insect pathogens S. marcescens (413) and P. luminescens (397), PhoP promotes transcription of the mgtE gene, which specifies a Mg2+ transporter of a different family (35). Although PhoP does not control the expression of CorA, the most widely distributed Mg2+ transporter in bacteria (35), it hinders CorA-mediated Mg2+ import by reversing membrane potential in uropathogenic E. coli (368) and, potentially, in other bacterial species.
In addition to promoting expression of Mg2+ importers, PhoP furthers cytoplasmic Mg2+ homeostasis by reducing the amounts of Mg2+-chelating ATP (146). Because ATP is the energy currency in all living cells, a reduction in ATP amounts has pleiotropic consequences for the bacterial proteome. For example, the PhoP decrease in ATP amounts provokes a systemic decrease in proteolysis of functional proteins by ATP-dependent proteases (414).
In S. Typhimurium, the PhoP-dependent ATP decrease is largely mediated by the MgtC protein (150), which is proposed to achieve this task by inhibiting the ATP-generating F1F0 ATPase under mildly acidic pH conditions (408). However, MgtC decreases ATP amounts in low Mg2+ even in organisms lacking F1F0 ATPase (240), and this decrease has recently been ascribed to MgtC’s ability to inhibit a phosphate uptake system (151). Despite lacking mgtC, E. coli also decreases ATP amounts when experiencing low cytoplasmic Mg2+ (38).
E. tarda lacks mgtC. However, the PhoP transcriptional activation of the F1F0 ATPase-specifying atpIBEFHAGDC operon (407) may also decrease ATP amounts. This is because the largest differences in mRNA abundance between wild-type and phoP strains is exhibited by the last four genes of the operon (407), which specify proteins that make up the F1 subunit of the ATPase. When expressed by itself, the F1 subunit can hydrolyze ATP but not make it (415). Therefore, PhoP/PhoQ appears to utilize different strategies to decrease ATP amounts among enteric species.
It has been hypothesized that the LPS constitutes a Mg2+ reservoir that Gram-negative species can access to satisfy the needs of cytoplasmic proteins that exhibit a strict dependence on Mg2+ (36, 127). Because Mg2+ neutralizes phosphates in the LPS, their covalent modification is critical to prevent electrostatic repulsion between adjacent LPS molecules. The modifications of the lipid A moiety of the LPS are under the control of the PhoP protein in many species.
In S. Typhimurium, PhoP controls the genes that mediate modification of lipid A phosphates with both L-Ara4N and phosphoethanolamine indirectly, via the PhoP-PmrD-PmrA pathway (127) (Fig. 5). By contrast, PhoP is a direct transcriptional activator of the genes responsible for the L-Ara4N modification of lipid A in Y. pestis, which lacks both pmrD, the gene mediating the connection between the PhoP/PhoQ and PmrA/PmrB systems in S. Typhimurium (40, 41), and pmrC, the gene responsible for modification of lipid A with phosphoethanolamine (232). K. pneumoniae activates the L-Ara4N modification genes both directly and indirectly, via the PhoP-PmrD-PmrA pathway (236). Thus, S. Typhimurium, Y. pestis, and K. pneumoniae share the qualitative property of modifying their lipid A phosphates with L-Ara4N in response to the low Mg2+ signal that activates the PhoP/PhoQ system. However, the three species differ in the kinetics of activation and deactivation of L-Ara4N modification genes (236, 416), reflecting the different network architectures used by these species to regulate these genes.
E. coli harbors the genes mediating L-Ara4N and phosphoethanolamine modification of lipid A. However, it fails to activate these genes when the inducing condition is low Mg2+ because its PmrD protein is highly divergent (173) and also because its PmrB protein has higher phosphatase activity towards PmrA-P than the PmrB protein from S. Typhimurium (170), and PmrD inhibits PmrB dephosphorylation of PmrA-P (40). E. coli can activate the L-Ara4N and phosphoethanolamine modification genes in a PhoP- and PmrD-dependent manner if the medium also contains Fe3+ (234, 235), which, as a PmrB-activating signal (417), likely decreases PmrB’s phosphatase activity toward PmrA-P, enabling a less active PmrD to protect PmrA-P.
By covalently modifying lipid A, L-Ara4N and phosphoethanolamine confer resistance to polymyxin B. Thus, organisms in which the corresponding modification genes are under PhoP control display polymyxin B resistance when experiencing PhoQ-inducing conditions. However, as discussed above, mildly acidic pH results in greater resistance than low Mg2+ (256) despite both signals activating PhoQ to a similar extent. This is because mildly acidic pH favors lipid A modification with both L-Ara4N and phosphoethanolamine, whereas low Mg2+ favors modification solely with L-Ara4N (256).
PhoP/PhoQ controls virulence in bacterial species that have very different lifestyles and host preferences. This is the case of the phytopathogens Pectobacterium and Dickeya (252, 399, 403); the zoopathogens Salmonella, Shigella, and Yersinia; and the insect pathogen Photorhabdus (397). The PhoP protein plays roles in both the mammalian and insect hosts that Y. pestis experiences during its infectious cycle (418). There appear to be some environment-specific functions controlled by PhoP/PhoQ, such as the alkalinization of the medium, that are a specific mechanism to attack plants (398). Similarly, the P. aeruginosa PhoP/PhoQ controls virulence by regulating the production of lipases and proteases, which then damage lung epithelial cells (409), unlike S. Typhimurium, where the phoP mutant is defective in intramacrophage survival (280). Curiously, the PhoP-activated mgtC gene contributes to virulence in both intracellular (158) and extracellular (419) pathogens.
Conservation of the Targets of PhoP Control
Despite the relatively good conservation in the signals acting on PhoP/PhoQ and functions controlled by PhoP/PhoQ, the PhoP regulon (i.e., the collection of genes regulated by PhoP) is far less conserved. This reflects a variety of genetic differences across bacterial species, including instances of promoter rewiring in which a gene and its homolog are present in two species that harbor PhoP, but PhoP controls the gene only in one of the species. For example, the MgtA protector protein MgtS is transcriptionally activated by PhoP in E. coli (59) but not in S. Typhimurium (169) due to differences in the corresponding promoter regions. Likewise, PhoP is a direct transcriptional activator of the rstA gene in S. Typhimurium but does not regulate the rstA homolog in Y. pestis (28). Conversely, PhoP promotes transcription of the psiE gene in Y. pestis but not in S. Typhimurium (28). Therefore, a regulatory interaction experimentally established between PhoP and a regulated gene in one species should not be readily extrapolated to a different species that also harbors PhoP and the regulated gene.
In addition, a large number of PhoP-regulated genes are horizontally acquired and thus exhibit a sporadic phylogenetic distribution among closely related bacterial species. For example, PhoP is a direct transcriptional activator of the ssrB, virK, mig-14, pagC, pcgL, pagK, and pipD genes, none of which are found in species outside the Salmonella genus. Similarly, PhoP regulates the horizontally acquired pelABCD, indA, and vfmE genes in Dickeya spp. (399). PhoP’s capacity to regulate horizontally acquired genes implies that the ancestral PhoP protein enables the display of species-specific properties.
PhoP’s ability to regulate horizontally acquired genes may reflect that foreign DNA is typically AT-rich and that the PhoP box is also AT-rich and remarkably conserved across bacterial species (10). By contrast, closely related bacterial species often differ in the promoter architecture of PhoP-activated horizontally acquired genes (46). For instance, S. Typhimurium and Y. pestis differ in the promoter architecture of their respective mgtC genes, which were independently acquired by these two species. This difference prevents the PhoP protein of one species from activating transcription of the mgtC promoter of the other species (46). This is in spite of the fact that both PhoP proteins exhibit wild-type binding to the PhoP boxes in the mgtC promoter regions of the two species (46). By contrast, the two PhoP proteins are indistinguishable in their ability to promote transcription of genes sharing promoter architecture.
Strains of a given species can differ in their phoP alleles, resulting in distinct phenotypic properties. For example, the phoP gene of modern virulent strains of Y. pestis differs from that in avirulent strains and from the Y. pestis ancestor Y. pseudotuberculosis at a single position. This change results in a single amino acid difference in the DNA-binding domain of the PhoP protein (172). Isogenic Y. pestis expressing a PhoP protein with a serine at position 215, which characterizes the virulent strains, expresses the PhoP-activated gene ugd in larger amounts than a strain with glycine at that position, which distinguishes avirulent Y. pestis (172). The ugd gene mediates the L-Ara-4N modification of the lipid A that results in polymyxin B resistance (127). Curiously, the PhoP S215-expressing strain is more resistant to polymyxin B than the PhoP G215-expressing strain at 21°C but similarly resistant at 37°C (172). These findings demonstrate how allelic differences in highly conserved genes can result in phenotypic differences within a given species. Moreover, they are reminiscent of S. enterica serovars that differ in a single amino acid in the DNA-binding domain of the related regulator PmrA (171).
CONCLUDING REMARKS
The PhoP/PhoQ system is a classical two-component system in which the sensor PhoQ responds to specific signals by changing the phosphorylated state of its cognate regulator PhoP, which, in turn, modifies bacterial behavior by altering gene expression. PhoP controls two main behaviors across bacterial species with different lifestyles: adaptation to low-Mg2+ conditions and pathogenicity. These behaviors are most often mediated by different genes in different species due to the pervasive impact that horizontal gene transfer and gene deletion have had on bacterial evolution. The investigation of the PhoP/PhoQ system has uncovered cellular responses and biochemical mechanisms later found in other regulatory systems and has enabled unbiased explorations of protein-protein interactions. The ongoing examination of the PhoP/PhoQ system will continue to reveal novel aspects of bacterial biology, signal transduction, and stress responses.
Biographies

Eduardo A. Groisman is the Waldemar Von Zedtwitz Professor of Microbial Pathogenesis at Yale School of Medicine and a member of the Microbial Sciences Institute at Yale University. He received an M.S. Biochemistry degree from the University of Buenos Aires, Argentina, and a Ph.D. from the University of Chicago, Illinois. His research program explores how a cell knows when, where, and for how long its gene products should be present. He addresses the various mechanisms raised by this question in bacteria that establish interactions with mammalians hosts.

Alexandre Duprey is a postdoctoral associate at the Groisman lab at Yale School of Medicine. He received an Engineering Degree from INSA Lyon, France, and then obtained his Ph.D. from Université de Lyon in 2016 under the supervision of Drs. William Nasser and Sylvie Reverchon. He seeks to understand and manipulate bacterial behaviors by acting on their regulatory networks and therefore, for the last 8 years, he has been studying the genetic mechanisms that underlie the behavior of pathogenic bacteria, both in plants (Dickeya) and in animals (Salmonella). He has a particular interest and expertise in the regulation of DNA supercoiling as well as its contributions to gene regulation in bacteria.

Jeongjoon Choi, Ph.D., received his bachelor’s degree from Seoul National University, South Korea. He earned his M.S. and Ph.D. in Food Molecular Microbiology at the same university under the supervision of Sangryeol Ryu, Ph.D., where he investigated how bacterial pathogens operate their virulence program. He continued his research at Yale School of Medicine with Eduardo A. Groisman, Ph.D., and he is currently an associate research scientist. He has investigated how bacterial pathogens modulate their behavior in response to environmental changes especially including those encountered during infection. He is particularly interested in one of such conditions, the mildly acidic pH that pathogens experience inside macrophage phagosome. He identified various biological processes enabling the pathogen Salmonella enterica to properly implement its virulence program in response to the mildly acidic pH during infection.
REFERENCES
Articles from Microbiology and Molecular Biology Reviews : MMBR are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/mmbr.00176-20
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8483708
Citations & impact
Impact metrics
Citations of article over time
Article citations
Constitutive activation of two-component systems reveals regulatory network interactions in Streptococcus agalactiae.
Nat Commun, 15(1):9175, 24 Oct 2024
Cited by: 1 article | PMID: 39448655 | PMCID: PMC11502775
Cytolysin A is an intracellularly induced and secreted cytotoxin of typhoidal Salmonella.
Nat Commun, 15(1):8414, 28 Sep 2024
Cited by: 0 articles | PMID: 39341826 | PMCID: PMC11438861
PhoPQ-mediated lipopolysaccharide modification governs intrinsic resistance to tetracycline and glycylcycline antibiotics in <i>Escherichia coli</i>.
mSystems, 9(10):e0096424, 30 Sep 2024
Cited by: 1 article | PMID: 39345149 | PMCID: PMC11495068
PhoP/PhoQ Two-Component System Contributes to Intestinal Inflammation Induced by Cronobacter sakazakii in Neonatal Mice.
Foods, 13(17):2808, 04 Sep 2024
Cited by: 0 articles | PMID: 39272573 | PMCID: PMC11394756
Multiple resistance factors collectively promote inoculum-dependent dynamic survival during antimicrobial peptide exposure in Enterobacter cloacae.
PLoS Pathog, 20(8):e1012488, 26 Aug 2024
Cited by: 0 articles | PMID: 39186812 | PMCID: PMC11379400
Go to all (46) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(2 citations)
PDBe - 3BQ8View structure
-
(1 citation)
PDBe - 1YAXView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Activation of master virulence regulator PhoP in acidic pH requires the Salmonella-specific protein UgtL.
Sci Signal, 10(494):eaan6284, 29 Aug 2017
Cited by: 30 articles | PMID: 28851823 | PMCID: PMC5966036
The PhoQ/PhoP regulatory network of Salmonella enterica.
Adv Exp Med Biol, 631:7-21, 01 Jan 2008
Cited by: 70 articles | PMID: 18792679
Review
Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system.
Mol Microbiol, 91(1):135-144, 20 Nov 2013
Cited by: 42 articles | PMID: 24256574 | PMCID: PMC3890429
Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon.
J Bacteriol, 177(15):4364-4371, 01 Aug 1995
Cited by: 179 articles | PMID: 7543474 | PMCID: PMC177185
Funding
Funders who supported this work.
HHS | NIH | National Institute of Allergy and Infectious Diseases (1)
Grant ID: AI49561
NIAID NIH HHS (1)
Grant ID: R01 AI049561
 a
,
b
a
,
b




