Abstract
Free full text

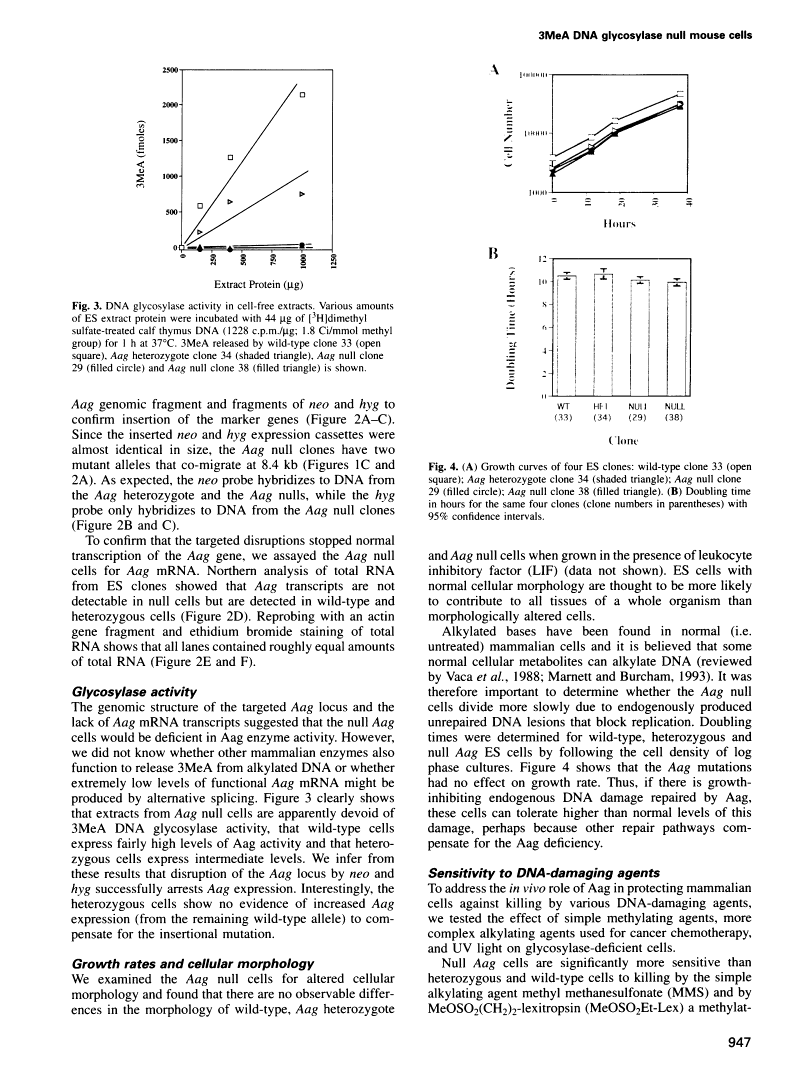
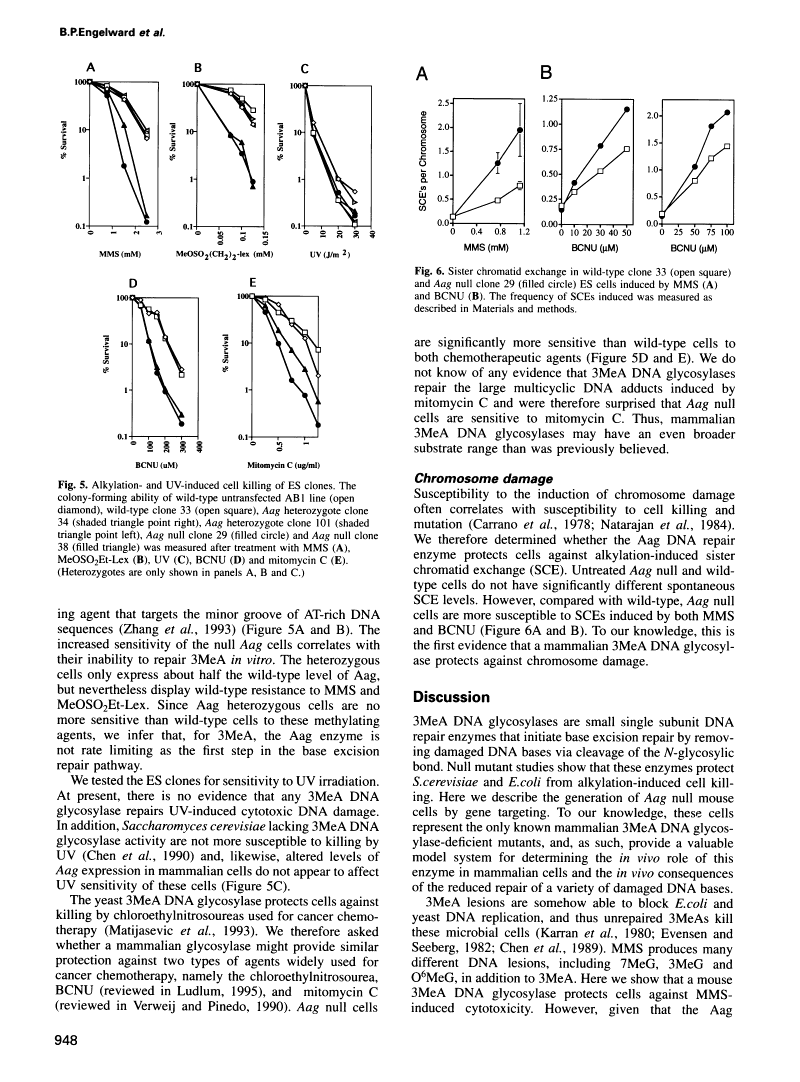
Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing.
Abstract
In Escherichia coli, the repair of 3-methyladenine (3MeA) DNA lesions prevents alkylation-induced cell death because unrepaired 3MeA blocks DNA replication. Whether this lesion is cytotoxic to mammalian cells has been difficult to establish in the absence of 3MeA repair-deficient cell lines. We previously isolated and characterized a mouse 3MeA DNA glycosylase cDNA (Aag) that provides resistance to killing by alkylating agents in E. coli. To determine the in vivo role of Aag, we cloned a large fragment of the Aag gene and used it to create Aag-deficient mouse cells by targeted homologous recombination. Aag null cells have no detectable Aag transcripts or 3MeA DNA glycosylase activity. The loss of Aag renders cells significantly more sensitive to methyl methanesulfonate-induced chromosome damage, and to cell killing induced by two methylating agents, one of which produces almost exclusively 3MeAs. Aag null embryonic stem cells become sensitive to two cancer chemotherapeutic alkylating agents, namely 1,3-bis(2-chloroethyl)-1-nitrosourea and mitomycin C, indicating that Aag status is an important determinant of cellular resistance to these agents. We conclude that this mammalian 3MeA DNA glycosylase plays a pivotal role in preventing alkylation-induced chromosome damage and cytotoxicity.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.7M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basu AK, Hanrahan CJ, Malia SA, Kumar S, Bizanek R, Tomasz M. Effect of site-specifically located mitomycin C-DNA monoadducts on in vitro DNA synthesis by DNA polymerases. Biochemistry. 1993 May 11;32(18):4708–4718. [Abstract] [Google Scholar]
- Berdal KG, Bjørås M, Bjelland S, Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990 Dec;9(13):4563–4568. [Europe PMC free article] [Abstract] [Google Scholar]
- Bessho T, Roy R, Yamamoto K, Kasai H, Nishimura S, Tano K, Mitra S. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8901–8904. [Europe PMC free article] [Abstract] [Google Scholar]
- Bjelland S, Bjørås M, Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993 May 11;21(9):2045–2049. [Europe PMC free article] [Abstract] [Google Scholar]
- Bjelland S, Birkeland NK, Benneche T, Volden G, Seeberg E. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J Biol Chem. 1994 Dec 2;269(48):30489–30495. [Abstract] [Google Scholar]
- Bodell WJ, Tokuda K, Ludlum DB. Differences in DNA alkylation products formed in sensitive and resistant human glioma cells treated with N-(2-chloroethyl)-N-nitrosourea. Cancer Res. 1988 Aug 15;48(16):4489–4492. [Abstract] [Google Scholar]
- Boiteux S, Huisman O, Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984 Nov;3(11):2569–2573. [Europe PMC free article] [Abstract] [Google Scholar]
- Carrano AV, Thompson LH, Lindl PA, Minkler JL. Sister chromatid exchange as an indicator of mutagenesis. Nature. 1978 Feb 9;271(5645):551–553. [Abstract] [Google Scholar]
- Chakravarti D, Ibeanu GC, Tano K, Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991 Aug 25;266(24):15710–15715. [Abstract] [Google Scholar]
- Chen J, Derfler B, Maskati A, Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen J, Derfler B, Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. [Europe PMC free article] [Abstract] [Google Scholar]
- Coulondre C, Miller JH. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. [Abstract] [Google Scholar]
- Dosanjh MK, Chenna A, Kim E, Fraenkel-Conrat H, Samson L, Singer B. All four known cyclic adducts formed in DNA by the vinyl chloride metabolite chloroacetaldehyde are released by a human DNA glycosylase. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):1024–1028. [Europe PMC free article] [Abstract] [Google Scholar]
- Dosanjh MK, Roy R, Mitra S, Singer B. 1,N6-ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase). Biochemistry. 1994 Feb 22;33(7):1624–1628. [Abstract] [Google Scholar]
- Engelward BP, Boosalis MS, Chen BJ, Deng Z, Siciliano MJ, Samson LD. Cloning and characterization of a mouse 3-methyladenine/7-methyl-guanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis. 1993 Feb;14(2):175–181. [Abstract] [Google Scholar]
- Erickson LC, Laurent G, Sharkey NA, Kohn KW. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumour cells. Nature. 1980 Dec 25;288(5792):727–729. [Abstract] [Google Scholar]
- Evensen G, Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. [Abstract] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993 Dec 3;75(5):1027–1038. [Abstract] [Google Scholar]
- Gonzaga PE, Brent TP. Affinity purification and characterization of human O6-alkylguanine-DNA alkyltransferase complexed with BCNU-treated, synthetic oligonucleotide. Nucleic Acids Res. 1989 Aug 25;17(16):6581–6590. [Europe PMC free article] [Abstract] [Google Scholar]
- Habraken Y, Ludlum DB. Release of chloroethyl ethyl sulfide-modified DNA bases by bacterial 3-methyladenine-DNA glycosylases I and II. Carcinogenesis. 1989 Mar;10(3):489–492. [Abstract] [Google Scholar]
- Habraken Y, Carter CA, Kirk MC, Ludlum DB. Release of 7-alkylguanines from N-(2-chloroethyl)-N'-cyclohexyl-N-nitrosourea-modified DNA by 3-methyladenine DNA glycosylase II. Cancer Res. 1991 Jan 15;51(2):499–503. [Abstract] [Google Scholar]
- Habraken Y, Carter CA, Sekiguchi M, Ludlum DB. Release of N2,3-ethanoguanine from haloethylnitrosourea-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Carcinogenesis. 1991 Oct;12(10):1971–1973. [Abstract] [Google Scholar]
- IYER VN, SZYBALSKI W. MITOMYCINS AND PORFIROMYCIN: CHEMICAL MECHANISM OF ACTIVATION AND CROSS-LINKING OF DNA. Science. 1964 Jul 3;145(3627):55–58. [Abstract] [Google Scholar]
- Karran P, Lindahl T, Ofsteng I, Evensen GB, Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. [Abstract] [Google Scholar]
- Klungland A, Fairbairn L, Watson AJ, Margison GP, Seeberg E. Expression of the E.coli 3-methyladenine DNA glycosylase I gene in mammalian cells reduces the toxic and mutagenic effects of methylating agents. EMBO J. 1992 Dec;11(12):4439–4444. [Europe PMC free article] [Abstract] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991 Aug 11;19(15):4293–4293. [Europe PMC free article] [Abstract] [Google Scholar]
- Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. [Abstract] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA, Nyström-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993 Dec 17;75(6):1215–1225. [Abstract] [Google Scholar]
- Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. [Abstract] [Google Scholar]
- Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. [Europe PMC free article] [Abstract] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988 Nov 24;336(6197):348–352. [Abstract] [Google Scholar]
- Marnett LJ, Burcham PC. Endogenous DNA adducts: potential and paradox. Chem Res Toxicol. 1993 Nov-Dec;6(6):771–785. [Abstract] [Google Scholar]
- Matijasevic Z, Sekiguchi M, Ludlum DB. Release of N2,3-ethenoguanine from chloroacetaldehyde-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9331–9334. [Europe PMC free article] [Abstract] [Google Scholar]
- Matijasevic Z, Boosalis M, Mackay W, Samson L, Ludlum DB. Protection against chloroethylnitrosourea cytotoxicity by eukaryotic 3-methyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11855–11859. [Europe PMC free article] [Abstract] [Google Scholar]
- McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984 Mar;3(3):545–550. [Europe PMC free article] [Abstract] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990 Sep 21;62(6):1073–1085. [Abstract] [Google Scholar]
- Mereau A, Grey L, Piquet-Pellorce C, Heath JK. Characterization of a binding protein for leukemia inhibitory factor localized in extracellular matrix. J Cell Biol. 1993 Aug;122(3):713–719. [Europe PMC free article] [Abstract] [Google Scholar]
- Natarajan AT, Simons JW, Vogel EW, van Zeeland AA. Relationship between cell killing, chromosomal aberrations, sister-chromatid exchanges and point mutations induced by monofunctional alkylating agents in Chinese hamster cells. A correlation with different ethylation products in DNA. Mutat Res. 1984 Aug;128(1):31–40. [Abstract] [Google Scholar]
- O'Connor TR, Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990 Oct;9(10):3337–3342. [Europe PMC free article] [Abstract] [Google Scholar]
- O'Connor TR, Laval J. Human cDNA expressing a functional DNA glycosylase excising 3-methyladenine and 7-methylguanine. Biochem Biophys Res Commun. 1991 May 15;176(3):1170–1177. [Abstract] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994 Mar 18;263(5153):1625–1629. [Abstract] [Google Scholar]
- Perry P, Wolff S. New Giemsa method for the differential staining of sister chromatids. Nature. 1974 Sep 13;251(5471):156–158. [Abstract] [Google Scholar]
- Popp RA, Stratton LP, Hawley DK, Effron K. Hemoglobin of mice with radiation-induced mutations at the hemoglobin loci. J Mol Biol. 1979 Jan 15;127(2):141–148. [Abstract] [Google Scholar]
- Ramírez-Solis R, Davis AC, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. [Abstract] [Google Scholar]
- Roy R, Brooks C, Mitra S. Purification and biochemical characterization of recombinant N-methylpurine-DNA glycosylase of the mouse. Biochemistry. 1994 Dec 20;33(50):15131–15140. [Abstract] [Google Scholar]
- Samson L, Linn S. DNA alkylation repair and the induction of cell death and sister chromatid exchange in human cells. Carcinogenesis. 1987 Feb;8(2):227–230. [Abstract] [Google Scholar]
- Samson L, Derfler B, Boosalis M, Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9127–9131. [Europe PMC free article] [Abstract] [Google Scholar]
- Santerre A, Britt AB. Cloning of a 3-methyladenine-DNA glycosylase from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2240–2244. [Europe PMC free article] [Abstract] [Google Scholar]
- Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):5873–5877. [Europe PMC free article] [Abstract] [Google Scholar]
- Singer B, Antoccia A, Basu AK, Dosanjh MK, Fraenkel-Conrat H, Gallagher PE, Kuśmierek JT, Qiu ZH, Rydberg B. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9386–9390. [Europe PMC free article] [Abstract] [Google Scholar]
- Snow ET, Foote RS, Mitra S. Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J Biol Chem. 1984 Jul 10;259(13):8095–8100. [Abstract] [Google Scholar]
- te Riele H, Maandag ER, Clarke A, Hooper M, Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990 Dec 13;348(6302):649–651. [Abstract] [Google Scholar]
- Thomas L, Yang CH, Goldthwait DA. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982 Mar 16;21(6):1162–1169. [Abstract] [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991 Jun 28;65(7):1153–1163. [Abstract] [Google Scholar]
- Vaca CE, Wilhelm J, Harms-Ringdahl M. Interaction of lipid peroxidation products with DNA. A review. Mutat Res. 1988 Mar;195(2):137–149. [Abstract] [Google Scholar]
- Vickers MA, Vyas P, Harris PC, Simmons DL, Higgs DR. Structure of the human 3-methyladenine DNA glycosylase gene and localization close to the 16p telomere. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3437–3441. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu ZN, Chan CL, Eastman A, Bresnick E. Expression of human O6-methylguanine-DNA methyltransferase in a DNA excision repair-deficient Chinese hamster ovary cell line and its response to certain alkylating agents. Cancer Res. 1992 Jan 1;52(1):32–35. [Abstract] [Google Scholar]
- Zhang Y, Chen FX, Mehta P, Gold B. Groove- and sequence-selective alkylation of DNA by sulfonate esters tethered to lexitropsins. Biochemistry. 1993 Aug 10;32(31):7954–7965. [Abstract] [Google Scholar]
Associated Data
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1002/j.1460-2075.1996.tb00429.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/j.1460-2075.1996.tb00429.x
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/j.1460-2075.1996.tb00429.x
Article citations
Transformed astrocytes confer temozolomide resistance on glioblastoma via delivering ALKBH7 to enhance APNG expression after educating by glioblastoma stem cells-derived exosomes.
CNS Neurosci Ther, 30(2):e14599, 01 Feb 2024
Cited by: 2 articles | PMID: 38332576 | PMCID: PMC10853646
DNA Alkylation Damage by Nitrosamines and Relevant DNA Repair Pathways.
Int J Mol Sci, 24(5):4684, 28 Feb 2023
Cited by: 8 articles | PMID: 36902118 | PMCID: PMC10003415
Review Free full text in Europe PMC
Mild phenotype of knockouts of the major apurinic/apyrimidinic endonuclease APEX1 in a non-cancer human cell line.
PLoS One, 16(9):e0257473, 16 Sep 2021
Cited by: 4 articles | PMID: 34529719 | PMCID: PMC8445474
Excision of mutagenic replication-blocking lesions suppresses cancer but promotes cytotoxicity and lethality in nitrosamine-exposed mice.
Cell Rep, 34(11):108864, 01 Mar 2021
Cited by: 11 articles | PMID: 33730582 | PMCID: PMC8527524
Focus on DNA Glycosylases-A Set of Tightly Regulated Enzymes with a High Potential as Anticancer Drug Targets.
Int J Mol Sci, 21(23):E9226, 03 Dec 2020
Cited by: 7 articles | PMID: 33287345 | PMCID: PMC7730500
Review Free full text in Europe PMC
Go to all (91) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cloning and characterization of a mouse 3-methyladenine/7-methyl-guanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11.
Carcinogenesis, 14(2):175-181, 01 Feb 1993
Cited by: 38 articles | PMID: 8435858
In vivo repair of methylation damage in Aag 3-methyladenine DNA glycosylase null mouse cells.
Nucleic Acids Res, 28(17):3294-3300, 01 Sep 2000
Cited by: 26 articles | PMID: 10954597 | PMCID: PMC110696
Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase.
Proc Natl Acad Sci U S A, 94(24):13087-13092, 01 Nov 1997
Cited by: 136 articles | PMID: 9371804 | PMCID: PMC24267
MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents.
DNA Repair (Amst), 6(8):1079-1099, 07 May 2007
Cited by: 369 articles | PMID: 17485253
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA055042
Grant ID: CA 55042
NIEHS NIH HHS (1)
Grant ID: P01-ES03926