Abstract
Free full text

Nobel Lecture
Prions
Abstract
Prions are unprecedented infectious pathogens that cause a group of invariably fatal neurodegenerative diseases by an entirely novel mechanism. Prion diseases may present as genetic, infectious, or sporadic disorders, all of which involve modification of the prion protein (PrP). Bovine spongiform encephalopathy (BSE), scrapie of sheep, and Creutzfeldt–Jakob disease (CJD) of humans are among the most notable prion diseases. Prions are transmissible particles that are devoid of nucleic acid and seem to be composed exclusively of a modified protein (PrPSc). The normal, cellular PrP (PrPC) is converted into PrPSc through a posttranslational process during which it acquires a high β-sheet content. The species of a particular prion is encoded by the sequence of the chromosomal PrP gene of the mammals in which it last replicated. In contrast to pathogens carrying a nucleic acid genome, prions appear to encipher strain-specific properties in the tertiary structure of PrPSc. Transgenetic studies argue that PrPSc acts as a template upon which PrPC is refolded into a nascent PrPSc molecule through a process facilitated by another protein. Miniprions generated in transgenic mice expressing PrP, in which nearly half of the residues were deleted, exhibit unique biological properties and should facilitate structural studies of PrPSc. While knowledge about prions has profound implications for studies of the structural plasticity of proteins, investigations of prion diseases suggest that new strategies for the prevention and treatment of these disorders may also find application in the more common degenerative diseases.
The torturous path of the scientific investigation that led to an understanding of familial Creutzfeldt–Jakob disease (CJD) chronicles a remarkable scientific odyssey. By 1930, the high incidence of familial (f) CJD in some families was known (1, 2). Almost 60 years were to pass before the significance of this finding could be appreciated (3–5). CJD remained a curious, rare neurodegenerative disease of unknown etiology throughout this period of three score years (6). Only with transmission of disease to apes after inoculation of brain extracts prepared from patients who died of CJD did the story begin to unravel (7).
Once CJD was shown to be an infectious disease, relatively little attention was paid to the familial form of the disease since most cases were not found in families. It is interesting to speculate how the course of scientific investigation might have proceeded had transmission studies not been performed until after the molecular genetic lesion had been identified. Had that sequence of events transpired, then the prion concept, which readily explains how a single disease can have a genetic or infectious etiology, might have been greeted with much less skepticism (8).
Epidemiologic studies designed to identify the source of the CJD infection were unable to identify any predisposing risk factors, although some geographic clusters were found (9–12). Libyan Jews living in Israel developed CJD about 30 times more frequently than other Israelis (13). This finding prompted some investigators to propose that the Libyan Jews had contracted CJD by eating lightly cooked brain from scrapie-infected sheep when they lived in Tripoli prior to emigration. Subsequently, the Libyan Jewish patients were all found to carry a mutation at codon 200 in their prion protein (PrP) gene (14–16).
My own interest in the subject began with a patient dying of CJD in the fall of 1972. At that time, I was beginning a residency in neurology and was most impressed by a disease process that could kill my patient in 2 months by destroying her brain while her body remained unaffected by this process. No febrile response, no leukocytosis or pleocytosis, no humoral immune response, and yet I was told that she was infected with a “slow virus.”
Slow Viruses.
The term “slow virus” had been coined by Bjorn Sigurdsson in 1954 while he was working in Iceland on scrapie and visna of sheep (17). Five years later, William Hadlow had suggested that kuru, a disease of New Guinea highlanders, was similar to scrapie and thus, it, too, was caused by a slow virus (18). Seven more years were to pass before the transmissibility of kuru was established by passaging the disease to chimpanzees inoculated intracerebrally (19). Just as Hadlow had made the intellectual leap between scrapie and kuru, Igor Klatzo made a similar connection between kuru and CJD (20). In both instances, these neuropathologists were struck by the similarities in light microscopic pathology of the central nervous system (CNS) that kuru exhibited with scrapie or CJD. In 1968, the transmission of CJD to chimpanzees after intracerebral inoculation was reported (7).
In scrapie, kuru, CJD, and all of the other disorders now referred to as prion diseases (Table (Table1), 1), spongiform degeneration and astrocytic gliosis is found upon microscopic examination of the CNS (Fig. (Fig.1)1) (21). The degree of spongiform degeneration is quite variable, whereas the extent of reactive gliosis correlates with the degree of neuron loss (22).
Table 1
The
prion diseases
diseases
| Disease | Host | Mechanism of pathogenesis |
|---|---|---|
| Kuru | Fore people | Infection through ritualistic cannibalism |
| iCJD | Humans | Infection from prion-contaminated HGH, dura mater grafts, etc. |
| vCJD | Humans | Infection from bovine prions? |
| fCJD | Humans | Germ-line mutations in PrP gene |
| GSS | Humans | Germ-line mutations in PrP gene |
| FFI | Humans | Germ-line mutation in PrP gene (D178N, M129) |
| sCJD | Humans | Somatic mutation or spontaneous conversion of PrPC into PrPSc? |
| FSI | Humans | Somatic mutation or spontaneous conversion of PrPC into PrPSc? |
| Scrapie | Sheep | Infection in genetically susceptible sheep |
| BSE | Cattle | Infection with prion-contaminated MBM |
| TME | Mink | Infection with prions from sheep or cattle |
| CWD | Mule deer, elk | Unknown |
| FSE | Cats | Infection with prion-contaminated bovine tissues or MBM |
| Exotic ungulate encephalopathy | Greater kudu, nyala, oryx | Infection with prion-contaminated MBM |
iCJD, iatrogenic CJD; vCJD, variant CJD; fCJD, familial CJD; sCJD, sporadic CJD; GSS, Gerstmann–Sträussler–Sheinker disease; FFI, fatal familial insomnia; FSI, fatal sporadic insomnia; BSE, bovine spongiform encephalopathy; TME, transmissible mink encephalopathy; CWD, chronic wasting disease; FSE, feline spongiform encephalopathy; HGH, human growth hormone; MBM, meat and bone meal.
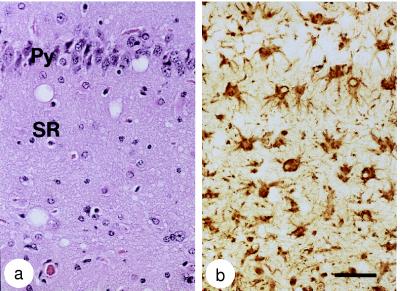
Neuropathologic changes in Swiss mice after inoculation with RML scrapie prions. (a) Hematoxylin and eosin stain of a serial section of the hippocampus shows spongiform degeneration of the neuropil, with vacuoles 10–30 μm in diameter. Brain tissue was immersion fixed in 10% buffered formalin solution after the animals had been sacrificed and was then embedded in paraffin. Py, pyramidal cell layer; SR, stratum radiatum. (b) Glial fibrillary acidic protein (GFAP) immunohistochemistry of a serial section of the hippocampus shows numerous reactive astrocytes. (Bar in b = 50 μm and also applies to a.) Photomicrographs were prepared by Stephen J. DeArmond.
Prions: A Brief Overview.
Before proceeding with a detailed discussion of our current understanding of prions causing scrapie and CJD, I provide a brief overview of prion biology. Prions are unprecedented infectious pathogens that cause a group of invariably fatal neurodegenerative diseases mediated by an entirely novel mechanism. Prion diseases may present as genetic, infectious, or sporadic disorders, all of which involve modification of the prion protein (PrP), a constituent of normal mammalian cells (23). CJD generally presents as progressive dementia, whereas scrapie of sheep and bovine spongiform encephalopathy (BSE) are generally manifest as ataxic illnesses (Table (Table1)1) (24).
Prions are devoid of nucleic acid and seem to be composed exclusively of a modified isoform of PrP designated PrPSc.‡ The normal, cellular PrP, denoted PrPC, is converted into PrPSc through a process whereby a portion of its α-helical and coil structure is refolded into β-sheet (25). This structural transition is accompanied by profound changes in the physicochemical properties of the PrP. The amino acid sequence of PrPSc corresponds to that encoded by the PrP gene of the mammalian host in which it last replicated. In contrast to pathogens with a nucleic acid genome that encode strain-specific properties in genes, prions encipher these properties in the tertiary structure of PrPSc (26–28). Transgenetic studies argue that PrPSc acts as a template upon which PrPC is refolded into a nascent PrPSc molecule through a process facilitated by another protein.
More than 20 mutations of the PrP gene are now known to cause the inherited human prion diseases, and significant genetic linkage has been established for five of these mutations (4, 16, 29–31). The prion concept readily explains how a disease can be manifest as a heritable as well as an infectious illness.
Resistance of Scrapie Agent to Radiation.
My fascination with CJD quickly shifted to scrapie once I learned of the remarkable radiobiological data that Tikvah Alper and her colleagues had collected on the scrapie agent (32–34). The scrapie agent had been found to be extremely resistant to inactivation by UV and ionizing radiation, as was later shown for the CJD agent (35). It seemed to me that the most intriguing question was the chemical nature of the scrapie agent; Alper’s data had evoked a torrent of hypotheses concerning its composition. Suggestions as to the nature of the scrapie agent ranged from small DNA viruses to membrane fragments to polysaccharides to proteins, the last of which eventually proved to be correct (36–42).
Scrapie of sheep and goats possesses a history no less fascinating than that of CJD. The resistance of the scrapie agent to inactivation by formalin and heat treatments (43), which were commonly used to produce vaccines against viral illnesses, suggested that the scrapie agent might be different from viruses, but it came at a time before the structure of viruses was understood. Later, this resistance was dismissed as an interesting observation but of little importance since some viruses can survive such treatments; indeed, this was not an unreasonable viewpoint. More than two decades were to pass before reports of the extreme resistance of the scrapie agent to inactivation by radiation again trumpeted the novelty of this infectious pathogen. Interestingly, British scientists had argued for many years about whether natural scrapie was a genetic or an infectious disease (44–46). Scrapie, like kuru and CJD, produced death of the host without any sign of an immune response to a “foreign infectious agent.”
My initial studies focused on the sedimentation properties of scrapie infectivity in mouse spleens and brains. From these studies, I concluded that hydrophobic interactions were responsible for the nonideal physical behavior of the scrapie particle (47, 48). Indeed, the scrapie agent presented a biochemical nightmare: infectivity was spread from one end to the other of a sucrose gradient and from the void volume to fractions eluting at 5–10 times the included volume of chromatographic columns. Such results demanded new approaches and better assays (49).
Bioassays.
As the number of hypotheses about the molecular nature of the scrapie agent began to exceed the number of laboratories working on this problem, the need for new experimental approaches became evident. Much of the available data on the properties of the scrapie agent had been gathered on brain homogenates prepared from mice with clinical signs of scrapie. These mice had been inoculated 4–5 months earlier with scrapie agent that originated in sheep but had been passaged multiple times in mice. Once an experiment was completed on these homogenates, an additional 12 months was required to obtain the results of an endpoint titration in mice (50). Typically, 60 mice were required to determine the titer of a single sample. This slow, tedious, and expensive system discouraged systematic investigation.
Although the transmission of scrapie to mice had ushered in a new era of research, the 1.5- to 2-year intervals between designing experiments and obtaining results discouraged sequential studies. Infrequently, the results of one set of experiments were used as a foundation for the next and so on. Moreover, the large number of mice needed to measure the infectivity in a single sample prevented studies where many experiments were performed in parallel. These problems encouraged publication of inconclusive experimental results.
In 1972, when I became fascinated by the enigmatic nature of the scrapie agent, I thought that the most direct path to determining the molecular structure of the scrapie agent was purification. Fortunately, I did not appreciate the magnitude of that task, although I had considerable experience and training in the purification of enzymes (51). Although many studies had been performed to probe the physicochemical nature of the scrapie agent by using the mouse endpoint titration system, few systematic investigations had been performed on the fundamental characteristics of the infectious scrapie particle (42). In fact, 12 years after introduction of the mouse bioassay, there were few data on the sedimentation behavior of the scrapie particle. Since differential centrifugation is frequently a useful initial step in the purification of many macromolecules, some knowledge of the sedimentation properties of the scrapie agent under defined conditions seemed mandatory. To perform such studies, Swiss mice were inoculated intracerebrally with the Chandler isolate of scrapie prions and the mice were sacrificed about 30 and 150 days later, when the titers in their spleens and brains, respectively, were expected to be at maximal levels. The two tissues were homogenized, extracted with detergent, and centrifuged for increasing times and speeds (47, 52). The disappearance of scrapie infectivity was measured in supernatant fractions by endpoint titration, which required 1 year to score.
Incubation time assays in hamsters.
In view of these daunting logistical problems, the identification of an inoculum that produced scrapie in the golden Syrian hamster (SHa) in ≈70 days after intracerebral inoculation proved to be an important advance (53, 54) once an incubation time assay was developed (55, 56). In earlier studies, SHa had been inoculated with prions, but serial passage with short incubation times was not reported (57). Development of the incubation time bioassay reduced the time required to measure prions in samples with high titers by a factor of 5: only 70 days were required instead of the 360 days previously needed. Equally important, 4 animals could be used in place of the 60 that were required for endpoint titrations, making possible a large number of parallel experiments. With this bioassay, research on the nature of the scrapie agent was accelerated nearly 100-fold and the hamster with high prion titers in its brain became the experimental animal of choice for biochemical studies.
The incubation time assay enabled development of effective purification schemes for enriching fractions for scrapie infectivity. It provided a means to assess quantitatively those fractions that were enriched for infectivity and those that were not. Such studies led rather rapidly to the development of a protocol for separating scrapie infectivity from most proteins and nucleic acids. With a ≈100-fold purification of infectivity relative to protein, >98% of the proteins and polynucleotides were eliminated, permitting more reliable probing of the constituents of these enriched fractions.
The Prion Concept.
As reproducible data began to accumulate indicating that scrapie infectivity could be reduced by procedures that hydrolyze or modify proteins but was resistant to procedures that alter nucleic acids, a family of hypotheses about the molecular architecture of the scrapie agent began to emerge (58). These data established, for the first time, that a particular macromolecule was required for infectivity and that this macromolecule was a protein. The experimental findings extended earlier observations on resistance of scrapie infectivity to UV irradiation at 250 nm (33) in that the four different procedures used to probe for a nucleic acid are based on physical principles that are independent of UV radiation damage.
Once the requirement of protein for infectivity was established, I thought that it was appropriate to give the infectious pathogen of scrapie a provisional name that would distinguish it from both viruses and viroids. After some contemplation, I suggested the term “prion,” derived from proteinaceous and infectious (58). At that time, I defined prions as proteinaceous infectious particles that resist inactivation by procedures that modify nucleic acids. I never imagined the irate reaction of some scientists to the word “prion”—it was truly remarkable!
Current definitions.
Perhaps the best current working definition of a prion is a proteinaceous infectious particle that lacks nucleic acid (28). Because a wealth of data supports the contention that scrapie prions are composed entirely of a protein that adopts an abnormal conformation, it is not unreasonable to define prions as infectious proteins (25, 27, 59, 60). But I hasten to add that we still cannot eliminate a small ligand bound to PrPSc as an essential component of the infectious prion particle. Learning how to renature PrPSc accompanied by restoration of prion infectivity or to generate prion infectivity de novo by using a synthetic polypeptide should help address this as-yet-unresolved issue (61). From a broader perspective, prions are elements that impart and propagate conformational variability.
Although PrPSc is the only known component of the infectious prion particles, these unique pathogens share several phenotypic traits with other infectious entities such as viruses. Because some features of the diseases caused by prions and viruses are similar, some scientists have difficulty accepting the existence of prions despite a wealth of scientific data supporting this concept (62–67).
Families of hypotheses.
Once the requirement for a protein was established, it was possible to revisit the long list of hypothetical structures that had been proposed for the scrapie agent and to eliminate carbohydrates, lipids, and nucleic acids as the infective elements within a scrapie agent devoid of protein (58). No longer could structures such as a viroid-like nucleic acid, a replicating polysaccharide, or a small polynucleotide surrounded by a carbohydrate be entertained as reasonable candidates to explain the puzzling properties of the scrapie agent (58, 68).
The family of hypotheses that remained after identifying a protein component was still large and required a continued consideration of all possibilities in which a protein was a critical element (49). The prion concept evolved from a family of hypotheses in which an infectious protein was only one of several possibilities. With the accumulation of experimental data on the molecular properties of the prion, it became possible to discard an increasing number of hypothetical structures. In prion research, as in many other areas of scientific investigation, a single hypothesis is all too often championed at the expense of a reasoned approach that requires entertaining a series of complex arguments until one or more can be discarded on the basis of experimental data (69).
Genes and DNA.
In some respects, the early development of the prion concept mirrors the story of DNA (70–72). Prior to the acceptance of DNA as the genetic material of life (73, 74), many scientists asserted that the DNA preparations must be contaminated with protein that is the true genetic material (75). The prejudices of these scientists were similar in some ways to those of investigators who have disputed the prion concept. But the scientists who attacked the hypothesis that genes are composed of DNA had no well proven alternative; they had only a set of feelings derived from poorly substantiated data sets that genes are made of protein. In contrast, those who attacked the hypothesis that the prion is composed only of protein had more than 30 years of cumulative evidence showing that genetic information in all organisms on our planet is encoded in DNA and that biological diversity resides in DNA. Studies of viruses and eventually viroids extended this concept to these small infectious pathogens (76) and showed that genes could also be composed of RNA (77, 78).
Discovery of the Prion Protein.
The discovery of the prion protein transformed research on scrapie and related diseases (79, 80). It provided a molecular marker that was subsequently shown to be specific for these illnesses as well as the major, and very likely the only, constituent of the infectious prion.
PrP 27–30 was discovered by enriching fractions from SHa brain for scrapie infectivity (79, 80). This protein is the protease-resistant core of PrPSc and has an apparent molecular mass of 27–30 kDa (81, 82). Although resistance to limited proteolysis proved to be a convenient tool for many but not all studies, use of proteases to enrich fractions for scrapie infectivity created a problem when the NH2-terminal sequence of PrP 27–30 was determined (81). The ragged NH2 terminus of PrP 27–30 yielded three sets of signals in almost every cycle of the Edman degradation. Only after these signals were properly interpreted and placed in correct register could a unique sequence be assigned for the NH2 terminus of PrP 27–30. The determination of the amino acid sequence of the NH2 terminus of PrP 27–30 made subsequent molecular cloning studies of the PrP gene possible (83, 84).
The finding that PrP mRNA levels were similar in normal uninfected and scrapie-infected tissues caused some investigators to argue that PrP 27–30 was not related to the infectious prion particle (83). An alternate interpretation prompted a search for a prion protein in uninfected animals that was found to be protease sensitive and soluble in nondenaturing detergents, unlike PrP 27–30. This isoform was designated PrPC (Fig. (Fig.2)2) (84, 85). Deduced amino acid sequences from PrP cDNA as well as immunoblotting studies revealed that PrP 27–30 was NH2-terminally truncated and was derived from a larger molecule, designated PrPSc, that was unique to infected animals (81, 82, 84–86).
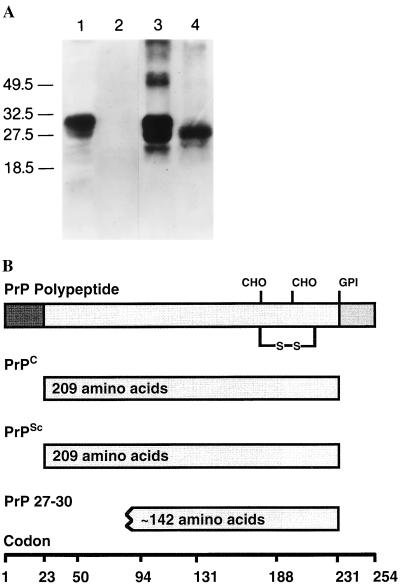
Prion protein isoforms. (A) Western immunoblot of brain homogenates from uninfected (lanes 1 and 2) and prion-infected (lanes 3 and 4) SHa. Samples in lanes 2 and 4 were digested with 50 μg/ml proteinase K for 30 min at 37°C. PrPC in lanes 2 and 4 was completely hydrolyzed under these conditions, whereas approximately 67 amino acids were digested from the NH2 terminus of PrPSc to generate PrP 27–30. After polyacrylamide gel electrophoresis (PAGE) and electrotransfer, the blot was developed with anti-PrP R073 polyclonal rabbit antiserum. Molecular size markers are in kilodaltons (kDa). (B) Bar diagram of SHaPrP, which consists of 254 amino acids. Attached carbohydrate (CHO) and a glycosyl-phosphatidylinositol (GPI) anchor are indicated. After processing of the NH2 and COOH termini, both PrPC and PrPSc consist of 209 residues. After limited proteolysis, the NH2 terminus of PrPSc is truncated to form PrP 27–30, which is composed of approximately 142 amino acids.
With the discovery of PrP 27–30 and the production of antiserum (87), brains from humans and animals with putative prion diseases were examined for the presence of this protein. In each case, PrP 27–30 was found, and it was absent in other neurodegenerative disorders such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (88–91). The extreme specificity of PrPSc for prion disease is an important feature of the protein and is consistent with the postulated role of PrPSc in both the transmission and pathogenesis of these illnesses (Table (Table2) 2) (92).
Table 2
Arguments for prions being composed largely, if not
entirely, of PrPSc molecules and devoid of
nucleic acid
acid
| 1 | PrPSc and scrapie infectivity copurify when biochemical and immunologic procedures are used. |
| 2 | The unusual properties of PrPSc mimic those of prions. Many different procedures that modify or hydrolyze PrPSc inactivate prions. |
| 3 | Levels of PrPSc are directly proportional to prion titers. Nondenatured PrPSc has not been separated from scrapie infectivity. |
| 4 | No evidence exists for a virus-like particle or a nucleic acid genome. |
| 5 | Accumulation of PrPSc is invariably associated with the pathology of prion diseases, including PrP amyloid plaques that are pathognomonic. |
| 6 | PrP gene mutations are genetically linked to inherited prion disease and cause formation of PrPSc. |
| 7 | Overexpression of PrPC increases the rate of PrPSc formation, which shortens the incubation time. Knock out of the PrP gene eliminates the substrate necessary for PrPSc formation and prevents both prion disease and prion replication. |
| 8 | Species variations in the PrP sequence are responsible, at least in part, for the species barrier that is found when prions are passaged from one host to another. |
| 9 | PrPSc preferentially binds to homologous PrPC, resulting in formation of nascent PrPSc and prion infectivity. |
| 10 | Chimeric and partially deleted PrP genes change susceptibility to prions from different species and support production of artificial prions with novel properties that are not found in nature. |
| 11 | Prion diversity is enciphered within the conformation of PrPSc. Strains can be generated by passage through hosts with different PrP genes. Prion strains are maintained by PrPC/PrPSc interactions. |
| 12 | Human prions from fCJD(E200K) and FFI patients impart different properties to chimeric MHu2M PrP in transgenic mice, which provides a mechanism for strain propagation. |
The accumulation of PrPSc contrasts markedly with that of glial fibrillary acidic protein (GFAP) in prion disease. In scrapie, GFAP mRNA and protein levels rise as the disease progresses (93), but the accumulation of GFAP is neither specific nor necessary for either the transmission or the pathogenesis of disease. Mice deficient for GFAP show no alteration in their incubation times (94, 95).
Except for PrPSc, no macromolecule has been found in tissues of patients dying of the prion diseases that is specific for these encephalopathies. In searches for a scrapie-specific nucleic acid, cDNAs have been identified that are complementary to mRNAs encoding other proteins with increased expression in prion disease (96–98). Yet none of the proteins has been found to be specific for prion disease.
Attempts to Falsify the Prion Hypothesis.
Numerous attempts to disprove the prion hypothesis over the past 15 years have failed. Such studies have tried unsuccessfully to separate scrapie infectivity from protein and more specifically from PrPSc. No preparations of purified prions containing less than one PrPSc molecule per ID50 unit have been reported (99), and no replication of prions in PrP-deficient (Prnp0/0) mice was found (100–104).
Copurification of PrP 27–30 and scrapie infectivity demands that the physicochemical properties as well as antigenicity of these two entities be similar (105) (Table (Table2).2). The results of a wide array of inactivation experiments demonstrated the similarities in the properties of PrP 27–30 and scrapie infectivity (61, 106–109). To explain these findings in terms of the virus hypothesis, it is necessary to postulate either a virus that has a coat protein which is highly homologous with PrP or a virus that binds tightly to PrPSc. In either case, the PrP-like coat protein or the PrPSc/virus complex must display properties indistinguishable from PrPSc alone. With each species that the putative virus invades, it must incorporate a new PrP sequence during replication.
Search for a scrapie-specific nucleic acid.
The inability to inactivate preparations highly enriched for scrapie infectivity by procedures that modify nucleic acids militates against the existence of a scrapie-specific nucleic acid (58, 110, 111). To explain the findings in terms of a virus, one must argue that PrPSc or an as-yet-undetected PrP-like protein of viral origin protects the viral genome from inactivation. The notion that the putative scrapie virus encodes a PrP-like protein was refuted by nucleic acid hybridization studies using a PrP cDNA probe. Less than 0.002 nucleic acid molecule encoding PrP per ID50 unit was found in purified preparations of SHa prions (84). To circumvent this finding, it could be hypothesized that the genetic code used by the PrP gene differs so greatly from that found in the cell that a PrP cDNA probe failed to detect it in highly purified preparations.
If prions contained a genome with a unique genetic code, then it is likely that this genome would encode some specialized proteins required for replication as well as some unique tRNAs. But both UV and ionizing radiation inactivation studies as well as physical studies have eliminated the possibility of a large nucleic acid hiding within purified preparations of prions (110–112). Only oligonucleotides of fewer than 50 bases were found at a concentration of one molecule per ID50 unit in prion preparations highly enriched for scrapie infectivity (113, 114). These small nucleic acids were of variable length and are thought to be degradation byproducts generated during purification of prions. Failure to find a bona fide genome was attributed to the unusual properties of the putative viral nucleic acid or technical incompetence on the part of the investigators who were unable to find it (63, 115).
PrP amyloid.
In preparations highly enriched for scrapie infectivity and containing only PrP 27–30 by silver staining of gels after SDS/PAGE, numerous rod-shaped particles were seen by electron microscopy after negative staining (Fig. (Fig.3)3) (107). Each of the rods was slightly different, in contrast to viruses, which exhibit extremely uniform structures (116). These irregular rods, composed largely, if not entirely, of PrP 27–30, were indistinguishable morphologically from many other purified amyloids (117). Studies of the prion rods with Congo red dye demonstrated that the rods also fulfilled the tinctorial criteria for amyloid (107), and immunostaining later showed that PrP is a major component of amyloid plaques in some animals and humans with prion disease (118–120). Subsequently, it was recognized that the prion rods were not required for scrapie infectivity (121); furthermore, the rods were shown to be an artifact of purification during which limited proteolysis of PrPSc generated PrP 27–30 that polymerized spontaneously in the presence of detergent (Fig. (Fig.3)3) (122).
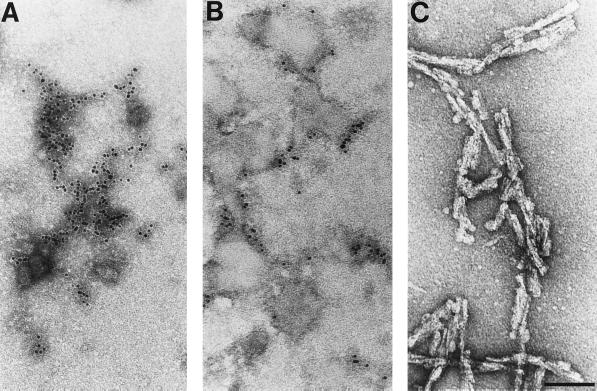
Electron micrographs of negatively stained and ImmunoGold-labeled prion proteins. (A) PrPC. (B) PrPSc. Neither PrPC nor PrPSc forms recognizable, ordered polymers. (C) Prion rods composed of PrP 27–30 were negatively stained. The prion rods are indistinguishable from many purified amyloids. (Bar = 100 nm.)
The idea that scrapie prions were composed of an amyloidogenic protein was truly heretical when it was introduced (107). Since the prevailing view at the time was that scrapie is caused by an atypical virus, many argued that amyloid proteins are mammalian polypeptides and not viral proteins!
Some investigators have argued that the prion rods are synonymous with scrapie-associated fibrils (123–125) even though morphologic and tinctorial features of these fibrils clearly differentiated them from amyloid and as such from the prion rods (126, 127). The scrapie-associated fibrils were identified by their unique ultrastructure in which two or four subfilaments were helically wound around each other (126) and were proposed to represent the first example of a filamentous animal virus (128). After the argument for a filamentous animal virus causing scrapie faded, it was hypothesized that a virus induces the formation of PrP amyloid to explain the accumulation of PrPSc in prion diseases (129). Besides the lack of evidence for a virus of any shape, no compelling data have been offered in support of the idea that prion diseases are caused by a filamentous bacterium called a spiroplasma (130).
Search for the ubiquitous “scrapie virus.”
When PrP gene mutations were discovered to cause familial prion diseases (4), it was postulated that PrPC is a receptor for the ubiquitous scrapie virus that binds more tightly to mutant than to wt PrPC (131). A similar hypothesis was proposed to explain why the length of the scrapie incubation time was found to be inversely proportional to the level of PrP expression in transgenic (Tg) mice and why Prnp0/0 mice are resistant to scrapie (132). The higher the level of PrP expression, the faster the spread of the putative virus, which results in shorter incubation times; conversely, mice deficient for PrP lack the receptor required for spread of the virus (63). The inability to find virus-like particles in purified preparations of PrPSc was attributed to these particles being hidden (115) even though tobacco mosaic viruses could be detected when one virion was added per ID50 unit of scrapie prions (121).
Recent studies on the transmission of mutant prions from FFI and fCJD(E200K) to Tg(MHu2M) mice, which results in the formation of two different PrPSc molecules (27), has forced a corollary to the ubiquitous virus postulate. To accommodate this result, at least two different viruses must reside worldwide, each of which binds to a different mutant HuPrPC and each of which induces a different MHu2M PrPSc conformer when transferred to Tg mice. Even more difficult to imagine is how one ubiquitous virus might acquire different mutant PrPSc molecules corresponding to FFI or fCJD(E200K) and then induce different MHu2M PrPSc conformers upon transmission to Tg mice.
Artificial prions.
To explain the production of artificial prions from chimeric or mutant PrP transgenes in terms of a virus (133–135), mutated PrPSc molecules must be incorporated into the virus. In the case of mice expressing chimeric PrP transgenes, artificial prions are produced with host ranges not previously found in nature. Similarly, deleting specific regions of PrP resulted in the formation of “miniprions” with a unique host range and neuropathology as described below. The production of artificial prions that were generated by modifying the PrP gene sequence and exhibit unique biological properties is another compelling argument against the proposition that scrapie and CJD are caused by viruses.
Skepticism once well justified.
While the skepticism about prions was once well justified and formed the basis for a vigorous scientific debate, the wealth of available data now renders such arguments moot. In summary, no single hypothesis involving a virus can explain the findings summarized above (Table (Table2);2); instead, a series of ad hoc hypotheses, virtually all of which can be refuted by experimental data, must be constructed to accommodate a steadily enlarging body of data.
It is notable that the search for an infectious pathogen with a nucleic acid genome as the cause of scrapie and CJD has done little to advance our understanding of these diseases. Instead, studies of PrP have created a wealth of data that now explain almost every aspect of these fascinating disorders. While no single experiment can refute the existence of the “scrapie virus,” all of the data taken together from numerous experimental studies present an impressive edifice which argues that the 50-year quest for a virus has failed because it does not exist!
Prions Defy Rules of Protein Structure.
Once cDNA probes for PrP became available, the PrP gene was found to be constitutively expressed in adult, uninfected brain (83, 84). This finding eliminated the possibility that PrPSc stimulated production of more of itself by initiating transcription of the PrP gene as proposed nearly two decades earlier (37). Determination of the structure of the PrP gene eliminated a second possible mechanism that might explain the appearance of PrPSc in brains already synthesizing PrPC. Since the entire protein coding region was contained within a single exon, there was no possibility for the two PrP isoforms to be the products of alternatively spliced mRNAs (82). Next, a posttranslational chemical modification that distinguishes PrPSc from PrPC was considered, but none was found in an exhaustive study (59) and we considered it likely that PrPC and PrPSc differed only in their conformation, a hypothesis also proposed earlier (37). However, this idea was no less heretical than that of an infectious protein.
For more than 25 years, it had been widely accepted that the amino acid sequence specifies one biologically active conformation of a protein (136). Yet in scrapie we were faced with the possibility that one primary structure for PrP might adopt at least two different conformations to explain the existence of both PrPC and PrPSc. When the secondary structures of the PrP isoforms were compared by optical spectroscopy, they were found to be markedly different (25). Fourier-transform infrared (FTIR) and circular dichroism (CD) studies showed that PrPC contains about 40% α-helix and little β-sheet, whereas PrPSc is composed of about 30% α-helix and 45% β-sheet (25, 137). Nevertheless, these two proteins have the same amino acid sequence!
Prior to comparative studies on the structures of PrPC and PrPSc, we found by metabolic labeling studies that the acquisition of PrPSc protease resistance is a posttranslational process (138). In our quest for a chemical difference that would distinguish PrPSc from PrPC, we found ethanolamine in hydrolysates of PrP 27–30, which signaled the possibility that PrP might contain a GPI anchor (139). Both PrP isoforms were found to carry GPI anchors, and PrPC was found on the surface of cells where it could be released by cleavage of the anchor. Subsequent studies showed that PrPSc formation occurs after PrPC reaches the cell surface (140, 141) and is localized to caveolae-like domains (142–145).
Modeling PrP structures.
Molecular modeling studies predicted that PrPC is a four-helix bundle protein containing four regions of secondary structure denoted H1, H2, H3, and H4 (Fig. (Fig.4)4) (146, 147). Subsequent NMR studies of a synthetic PrP peptide containing residues 90–145 provided good evidence for H1 (148). This peptide contains the residues 113–128, which are most highly conserved among all species studied (Fig. (Fig.44A) (147, 149, 150) and correspond to a transmembrane region of PrP that was delineated in cell-free translation studies (151, 152). Recent studies show that a transmembrane form of PrP accumulates in GSS caused by the A117V mutation and in Tg mice overexpressing either mutant or wild-type (wt)PrP (153). The paradoxical lack of evidence for an α-helix in this region from NMR studies of recombinant PrP in aqueous buffers (154–156) could be explained if the recombinant PrPs correspond to the secreted form of PrP that was also identified in cell-free translation studies. This contention is supported by studies with recombinant antibody fragments (Fabs) showing the GPI-anchored PrPC on the surface of cells exhibits an immunoreactivity similar to that of recombinant PrP that was prepared with an α-helical conformation (157, 158). GPI-anchored PrPC is synthesized within the secretory pathway and transported to the surface of the cell (139, 159).
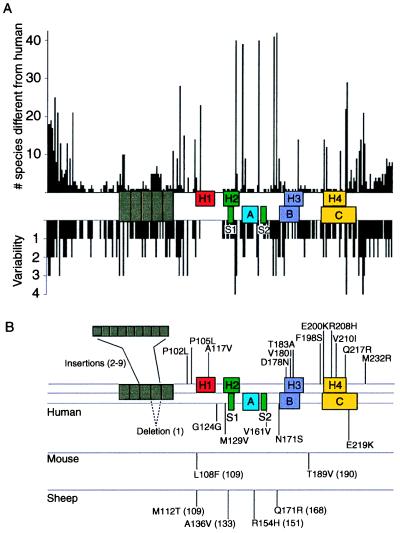
Species variations and mutations of the prion protein gene. (A) Species variations. The x-axis represents the human PrP sequence, with the five octarepeats and H1–H4 regions of putative secondary structure shown as well as the three α-helices A, B, and C and the two β-strands S1 and S2 as determined by NMR. The precise residues corresponding to each region of secondary structure are given in Fig. Fig.5.5. Vertical bars above the axis indicate the number of species that differ from the human sequence at each position. Below the axis, the length of the bars indicates the number of alternative amino acids at each position in the alignment. (B) Mutations causing inherited human prion disease and polymorphisms in human, mouse, and sheep. Above the line of the human sequence are mutations that cause prion disease. Below the lines are polymorphisms, some but not all of which are known to influence the onset as well as the phenotype of disease. Data were compiled by Paul Bamborough and Fred E. Cohen.
Optical spectroscopic measurements of PrPC provided the necessary background for more detailed structural studies (25). Unable to produce crystals of PrP, we and others utilized NMR to determine the structure of an α-helical form of a recombinant PrP. The NMR structure of a COOH-terminal fragment of MoPrP consisting of 111 residues showed three helices, two of which corresponded to H3 and H4 in the PrPC model, and two small β-strands each consisting of three residues (154). How the structure of this MoPrP(121–231) fragment differs from PrPC is of interest because this fragment is lethal when expressed in Tg mice (160). Subsequently, structural studies were performed on a longer fragment of PrP containing residues 90–231 and corresponding to SHaPrP 27–30 (155, 161, 162). Expression of PrP(90–231) in Tg mice did not produce spontaneous disease (163, 164). More recently, NMR structures of recombinant full-length PrP have been reported (156, 165).
Models of PrPSc suggested that formation of the disease-causing isoform involves refolding of residues within the region between residues 90 and 140 into β-sheets (166); the single disulfide bond joining COOH-terminal helices would remain intact because the disulfide is required for PrPSc formation (Fig. (Fig.55E) (167, 168). The high β-sheet content of PrPSc was predicted from the ability of PrP 27–30 to polymerize into amyloid fibrils (107). Subsequent optical spectroscopy confirmed the presence of β-sheet in both PrPSc and PrP 27–30 (25, 169–171). Deletion of each of the regions of putative secondary structure in PrP, except for the NH2-terminal 66 amino acids (residues 23–88) (163, 172) and a 36-amino acid region (mouse residues 141–176) prevented formation of PrPSc as measured in scrapie–infected cultured neuroblastoma cells (168). With anti-PrP Fabs selected from phage display libraries (157) and two monoclonal antibodies (mAbs) derived from hybridomas (173–175), the major conformational change that occurs during conversion of PrPC into PrPSc has been localized largely, but not entirely, to a region bounded by residues 90 and 112 (158). Similar conclusions were drawn from studies with an anti-PrP IgM mAb (176). While these results indicate that PrPSc formation involves primarily a conformational change in the domain composed of residues 90–112, mutations causing inherited prion diseases have been found throughout the protein (Fig. (Fig.44B). Interestingly, all of the known point mutations in PrP with biological significance occur either within or adjacent to regions of putative secondary structure in PrP and as such, appear to destabilize the structure of PrP (147, 148, 154).
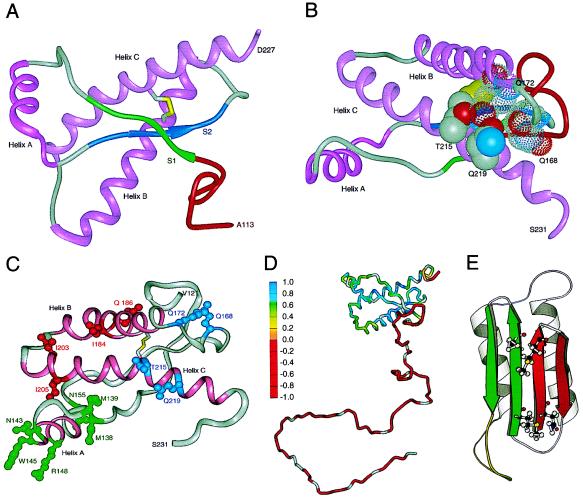
Structures of prion proteins. (A) NMR structure of SHa recombinant (r) PrP(90–231). Presumably, the structure of the α-helical form of rPrP(90–231) resembles that of PrPC. rPrP(90–231) is viewed from the interface where PrPSc is thought to bind to PrPC. The color scheme is as follows: α-helices A (residues 144–157), B (172–193), and C (200–227) in pink; disulfide between Cys-179 and Cys-214 in yellow; conserved hydrophobic region composed of residues 113–126 in red; loops in gray; residues 129–134 in green encompassing strand S1 and residues 159–165 in blue encompassing strand S2; the arrows span residues 129–131 and 161–163, as these show a closer resemblance to β-sheet (155). (B) NMR structure of rPrP(90–231) is viewed from the interface where protein X is thought to bind to PrPC. Protein X appears to bind to the side chains of residues that form a discontinuous epitope: some amino acids are in the loop composed of residues 165–171 and at the end of helix B (Gln-168 and Gln-172 with a low-density van der Waals rendering), whereas others are on the surface of helix C (Thr-215 and Gln-219 with a high-density van der Waals rendering) (178). (C) PrP residues governing the transmission of prions (180). NMR structure of recombinant SHaPrP region 121–231 (155) shown with the putative epitope formed by residues 184, 186, 203, and 205 highlighted in red. Residue numbers correspond to SHaPrP. Additional residues (138, 139, 143, 145, 148, and 155) that might participate in controlling the transmission of prions across species are depicted in green. Residues 168, 172, 215, and 219 that form the epitope for the binding of protein X are shown in blue. The three helices (A, B, and C) are highlighted in pink. (D) Schematic diagram showing the flexibility of the polypeptide chain for PrP(29–231) (156). The structure of the portion of the protein representing residues 90–231 was taken from the coordinates of PrP(90–231) (155). The remainder of the sequence was hand-built for illustration purposes only. The color scale corresponds to the heteronuclear {1H}-15N nuclear Overhauser enhancement data: red for the lowest (most negative) values, where the polypeptide is most flexible, to blue for the highest (most positive) values in the most structured and rigid regions of the protein. (E) Plausible model for the tertiary structure of HuPrPSc (166). Color scheme is as follows: S1 β-strands are 108–113 and 116–122 in red; S2 β-strands are 128–135 and 138–144 in green; α-helices H3 (residues 178–191) and H4 (residues 202–218) in gray; loop (residues 142–177) in yellow. Four residues implicated in the species barrier are shown in ball-and-stick form (Asn-108, Met-112, Met-129, Ala-133).
NMR structure of recombinant PrP.
The NMR structure of recombinant (r) SHaPrP(90–231) derived from Escherichia coli was determined after the protein was purified and refolded (Fig. (Fig.55A). Residues 90–112 are not shown because marked conformational heterogeneity was found in this region, while residues 113–126 constitute the conserved hydrophobic region, which also displays some structural plasticity (162). Although some features of the structure of rPrP(90–231) are similar to those reported earlier for the smaller recombinant MoPrP(121–231) fragment (154, 177), substantial differences were found. For example, the loop at the NH2 terminus of helix B is well defined in rPrP(90–231) but is disordered in MoPrP(121–231); in addition, helix C is composed of residues 200–227 in rPrP(90–231) but extends only from 200–217 in MoPrP(121–231). The loop and the COOH-terminal portion of helix C are particularly important because they form the site to which protein X binds as described below (Fig. (Fig.55B) (178). Whether the differences between the two recombinant PrP fragments are because of (i) their different lengths, (ii) species-specific differences in sequences, or (iii) the conditions used for solving the structures remains to be determined.
Studies of chimeric SHa/Mo and Hu/Mo PrP transgenes identified a domain composed of residues 95–170, where PrPC binds to PrPSc (133, 179). When chimeric bovine (Bo)/Mo PrP transgenes failed to render mice sensitive to BSE prions, we examined the differences among the sequences in the chimeric and parent PrP genes (180). The findings identified a second domain in PrP composed of residues 180–205 that seems to modulate the interaction between PrPC and PrPSc (Fig. (Fig.55C).
Recent NMR studies of full-length MoPrP(23–231) and SHaPrP(29–231) have shown that the NH2 termini are highly flexible and lack identifiable secondary structure under the experimental conditions employed (Fig. (Fig.55D) (156, 165). Studies of SHaPrP(29–231) indicate transient interactions between the COOH terminus of helix B and the highly flexible, NH2-terminal random-coil containing the octareapeats (residues 29–125) (156); such interactions were not reported for MoPrP(23–231) (165).
PrP appears to bind copper.
The highly flexible NH2 terminus of recombinant PrP may be more structured in the presence of copper. Each SHaPrP(29–231) molecule was found to bind two atoms of Cu2+; other divalent cations did not bind to PrP (181). Earlier studies with synthetic peptides corresponding to the octarepeat sequence demonstrated the binding of Cu2+ ions (182, 183), and optical spectroscopy showed that Cu2+ induced an α-helix formation in these peptides (184). More recently, PrP-deficient (Prnp0/0) mice were found to have lower levels of Zn/Cu superoxide dismutase (SOD) activity than do controls (185); SOD activity has been shown to mirror the state of copper metabolism (186). Measurements of membrane extracts from brains of Prnp0/0 mice showed low levels of Cu, whereas Fe and Zn were unchanged suggesting PrPC might function as a Cu2+-binding protein (187).
Disturbances in Cu2+ homeostasis leading to dysfunction of the CNS are well documented in humans and animals but are not known to be due to abnormalities in PrP metabolism: Menkes disease is manifest at birth and is due to a mutation of the MNK gene on the X chromosome, whereas Wilson’s disease appears in childhood and is due to a mutation of the WD gene on chromosome 13 (188–191). Both the MNK and WD genes encode copper-transporting ATPases. While both Menkes and Wilson’s diseases are recessive disorders, only Wilson’s disease can be treated with copper-chelating reagents. Interestingly, cuprizone, a Cu2+-chelating reagent, has been used in mice to induce neuropathological changes similar to those found in the prion diseases (192, 193).
PrP Gene Structure and Expression.
The entire open reading frame (ORF) of all known mammalian and avian PrP genes resides within a single exon (4, 82, 194, 195). The mouse, sheep, cattle, and rat PrP genes contain three exons with the ORFs in exon 3 (196–200) which is analogous to exon 2 of the SHa gene (82). The two exons of the SHaPrP gene are separated by a 10-kb intron: exon 1 includes a portion of the 5′ untranslated leader sequence, while exon 2 includes the ORF and 3′ untranslated region (82). Recently, a low abundance SHaPrP mRNA containing an additional small exon in the 5′ untranslated region was discovered that is encoded by the SHaPrP gene (201). Comparative sequencing of sheep and Hu cosmid clones containing PrP genes revealed an additional putative, small untranslated 5′ exon in the HuPrP gene (202). The promoters of both the SHa- and MoPrP genes contain multiple copies of G+C-rich repeats and are devoid of TATA boxes. These G+C nonamers represent a motif that may function as a canonical binding site for the transcription factor Sp1 (203). Mapping of PrP genes to the short arm of Hu chromosome 20 and to the homologous region of Mo chromosome 2 argues for the existence of PrP genes prior to the speciation of mammals (204, 205).
Although PrP mRNA is constitutively expressed in the brains of adult animals (83, 84), it is highly regulated during development. In the septum, levels of PrP mRNA and choline acetyltransferase were found to increase in parallel during development (206). In other brain regions, PrP gene expression occurred at an earlier age. In situ hybridization studies show that the highest levels of PrP mRNA are found in neurons (207).
PrPC expression in brain was defined by standard immunohistochemistry (208) and by histoblotting in the brains of uninfected controls (209). Immunostaining of PrPC in the SHa brain was most intense in the stratum radiatum and stratum oriens of the CA1 region of the hippocampus and was virtually absent from the granule cell layer of the dentate gyrus and the pyramidal cell layer throughout Ammon’s horn. PrPSc staining was minimal in those regions that were intensely stained for PrPC. A similar relationship between PrPC and PrPSc was found in the amygdala. In contrast, PrPSc accumulated in the medial habenular nucleus, the medial septal nuclei, and the diagonal band of Broca; in contrast, these areas were virtually devoid of PrPC. In the white matter, bundles of myelinated axons contained PrPSc but were devoid of PrPC. These findings suggest that prions are transported along axons and are in agreement with earlier findings in which scrapie infectivity was found to migrate in a pattern consistent with retrograde transport (210–212).
Molecular Genetics of Prion Diseases.
Independent of enriching brain fractions for scrapie infectivity that led to the discovery of PrPSc, the PrP gene was shown to be genetically linked to a locus controlling scrapie incubation times (213). Subsequently, mutation of the PrP gene was shown to be genetically linked to the development of familial prion disease (4). At the same time, expression of a SHaPrP transgene in mice was shown to render the animals highly susceptible to SHa prions, demonstrating that expression of a foreign PrP gene could abrogate the species barrier (214). Later, PrP-deficient (Prnp0/0) mice were found to be resistant to prion infection and failed to replicate prions, as expected (100, 101). The results of these studies indicated PrP must play a central role in the transmission and pathogenesis of prion disease, but equally important, they argued that the abnormal isoform is an essential component of the prion particle (23).
PrP gene dosage controls length of incubation time.
Scrapie incubation times in mice were used to distinguish prion strains and to identify a gene controlling its length (135, 215). This gene was initially called Sinc on the basis of genetic crosses between C57BL and VM mice that exhibited short and long incubation times, respectively (215). Because of the restricted distribution of VM mice, we searched for another mouse with long incubation times. I/Ln mice proved to be a suitable substitute for VM mice (216); eventually, I/Ln and VM mice were found to be derived from a common ancestor (217). With a PrP cDNA probe, we demonstrated genetic linkage between the PrP gene and a locus controlling the incubation time in crosses between NZW/LacJ and I/Ln mice (213). We provisionally designated these genes as components of the Prn complex but eventually found that the incubation time gene, Prn-i, is either congruent with or closely linked to the PrP gene, Prnp (195).
Although the amino acid substitutions in PrP that distinguish NZW (Prnpa ) from I/Ln (Prnpb) mice argued for congruency of Prnp and Prn-i, experiments with Prnpa mice expressing Prnpb transgenes demonstrated a “paradoxical” shortening of incubation times (196). We had predicted that these Tg mice would exhibit a prolongation of the incubation time after inoculation with RML prions on the basis of (Prnpa × Prnpb)F1 mice, which do exhibit long incubation times. We described those findings as “paradoxical shortening” because we and others had believed for many years that long incubation times are dominant traits (213, 215). From studies of congenic and transgenic mice expressing different numbers of the a and b alleles of Prnp, we learned that these findings were not paradoxical; indeed, they resulted from increased PrP gene dosage (218). When the RML isolate was inoculated into congenic and transgenic mice, increasing the number of copies of the a allele was found to be the major determinant in reducing the incubation time; however, increasing the number of copies of the b allele also reduced the incubation time, but not to the same extent as that seen with the a allele. From the foregoing investigations, we concluded that both Sinc and Prn-i are congruent with PrP (218), and recent gene targeting studies have confirmed this view (219).
Overexpression of wtPrP transgenes.
Mice were constructed expressing different levels of the wt SHaPrP transgene (214). Inoculation of these Tg(SHaPrP) mice with SHa prions demonstrated abrogation of the species barrier, resulting in abbreviated incubation times (220). The length of the incubation time after inoculation with SHa prions was inversely proportional to the level of SHaPrPC in the brains of Tg(SHaPrP) mice (220). Bioassays of brain extracts from clinically ill Tg(SHaPrP) mice inoculated with Mo prions revealed that only Mo prions but no SHa prions were produced. Conversely, inoculation of Tg(SHaPrP) mice with SHa prions led only to the synthesis of SHa prions. Although the rate of PrPSc synthesis appears to be a function of the level of PrPC expression in Tg mice, the level to which PrPSc finally accumulates seems to be independent of PrPC concentration (220).
During transgenetic studies, we discovered that uninoculated older mice harboring numerous copies of wtPrP transgenes derived from Syrian hamsters, sheep, and Prnpb mice spontaneously developed truncal ataxia, hind-limb paralysis, and tremors (198). These Tg mice exhibited a profound necrotizing myopathy involving skeletal muscle, a demyelinating polyneuropathy, and focal vacuolation of the CNS. Development of disease was dependent on transgene dosage. For example, Tg(SHaPrP+/+)7 mice homozygous for the SHaPrP transgene array regularly developed disease between 400 and 600 days of age, whereas hemizygous Tg(SHaPrP+/0)7 mice also developed disease, but only after >650 days.
PrP-deficient mice.
The development and lifespan of two lines of PrP-deficient (Prnp0/0) mice were indistinguishable from those of controls (221, 222), whereas two other lines exhibited ataxia and Purkinje cell degeneration at ≈70 weeks of age (223) (R. Moore and D. Melton, personal communication). In the former two lines with normal development, altered sleep–wake cycles have been reported (224), and altered synaptic behavior in brain slices was reported (225, 226) but could not be confirmed by others (227, 228).
Prnp0/0 mice are resistant to prions (100, 101). Prnp0/0 mice were sacrificed 5, 60, 120, and 315 days after inoculation with RML prions, and brain extracts were bioassayed in CD-1 Swiss mice. Except for residual infectivity from the inoculum detected at 5 days after inoculation, no infectivity was detected in the brains of Prnp0/0 mice (101). One group of investigators found that Prnp0/0 mice inoculated with RML prions and sacrificed 20 weeks later had 103.6 ID50 units/ml of homogenate by bioassay (100). Others have used this report to argue that prion infectivity replicates in the absence of PrP (67, 132). Neither we nor the authors of the initial report could confirm the finding of prion replication in Prnp0/0 mice (101, 103).
Prion Species Barrier and Protein X.
The passage of prions between species is often a stochastic process, almost always characterized by prolonged incubation times during the first passage in the new host (36). This prolongation is often referred to as the “species barrier” (36, 229). Prions synthesized de novo reflect the sequence of the host PrP gene and not that of the PrPSc molecules in the inoculum derived from the donor (90). On subsequent passage in a homologous host, the incubation time shortens to that recorded for all subsequent passages. From studies with Tg mice, three factors have been identified that contribute to the species barrier: (i) the difference in PrP sequences between the prion donor and recipient, (ii) the strain of prion, and (iii) the species specificity of protein X, a factor defined by molecular genetic studies that binds to PrPC and facilitates PrPSc formation. This factor is likely to be a protein, hence the provisional designation protein X (134, 178). The prion donor is the last mammal in which the prion was passaged and its PrP sequence represents the “species” of the prion. The strain of prion, which seems to be enciphered in the conformation of PrPSc, conspires with the PrP sequence, which is specified by the recipient, to determine the tertiary structure of nascent PrPSc. These principles are demonstrated by studies on the transmission of SHa prions to mice showing that expression of a SHaPrP transgene in mice abrogated the species barrier (Table (Table3)3) (214). Besides the PrP sequence, the strain of prion modified transmission of SHa prions to mice (Table (Table3)3) (135, 230, 231).
Table 3
Influence of prion species and strains on transmission
across a
species barrier
barrier
| Inoculum | Host | Incubation time, days
(n/n0)
| |
|---|---|---|---|
| Sc237 | 139H | ||
| SHa | SHa | 77 ± ± 1
1 (48/48) (48/48) | 167 ± ± 1 1 (94/94) (94/94) |
| SHa | Non-Tg mice | >700 (0/9) (0/9) | 499 ± ± 15 15 (11/11) (11/11)
|
| SHa | Tg(SHaPrP)81/FVB mice | 75 ± ± 2 2 (22/22) (22/22) | 110 ±
± 2 2 (19/19) (19/19)
|
| SHa | Tg(SHaPrP)81/Prnp0/0 mice | 54 ±
± 1 1 (9/9) (9/9) | 65 ± ± 1 1 (15/15) (15/15) |
Transmission of Hu prions.
Protein X was postulated to explain the results on the transmission of Hu prions to Tg mice (Table (Table4)4) (134, 179). Mice expressing both Mo and HuPrP were resistant to Hu prions, whereas those expressing only HuPrP were susceptible. These results argue that MoPrPC inhibited transmission of Hu prions—i.e., the formation of nascent HuPrPSc. In contrast to the foregoing studies, mice expressing both MoPrP and chimeric MHu2MPrP were susceptible to Hu prions and mice expressing MHu2MPrP alone were only slightly more susceptible. These findings contend that MoPrPC has only a minimal effect on the formation of chimeric MHu2MPrPSc.
Table 4
Evidence for protein X from transmission studies of
human prions
prions
| Inoculum | Host | MoPrP gene | Incubation time, days | (n/n0) |
|---|---|---|---|---|
| sCJD | Tg(HuPrP) | Prnp+/+ | 721 | (1/10) |
| sCJD | Tg(HuPrP) | Prnp0/0 | 263 ±
± 2 2 | (6/6) |
| sCJD | Tg(MHu2M) | Prnp+/+ | 238 ± ± 3
3 | (8/8) |
| sCJD | Tg(MHu2M) | Prnp0/0 | 191 ± ± 3
3 | (10/10) |
Data with inoculum RG (134); incubation times are given ±SEM.
Genetic evidence for protein X.
When the data on Hu prion transmission to Tg mice were considered together, they suggested that MoPrPC prevented the conversion of HuPrPC into PrPSc but had little effect on the conversion of MHu2M into PrPSc by binding to another Mo protein. We interpreted these results in terms of MoPrPC binding to this Mo protein with a higher affinity than does HuPrPC. We postulated that MoPrPC had little effect on the formation of PrPSc from MHu2M (Table (Table4)4) because MoPrP and MHu2M share the same amino acid sequence at the COOH terminus. We hypothesized that MoPrPC only weakly inhibited transmission of SHa prions to Tg(SHaPrP) mice (Table (Table3)3) because SHaPrP is more closely related to MoPrP than is HuPrP.
Using scrapie-infected Mo neuroblastoma cells transfected with chimeric Hu/Mo PrP genes, we extended our studies of protein X. Substitution of a Hu residue at position 214 or 218 prevented PrPSc formation (Fig. (Fig.5B)5B) (178). The side chains of these residues protrude from the same surface of the COOH-terminal α-helix, forming a discontinuous epitope with residues 167 and 171 in an adjacent loop. Substitution of a basic residue at position 167, 171, or 218 prevented PrPSc formation; these mutant PrPs appear to act as “dominant negatives” by binding protein X and rendering it unavailable for prion propagation. Our findings within the context of protein X explain the protective effects of basic polymorphic residues in PrP of humans and sheep (199, 232, 233).
Is protein X a molecular chaperone?
Since PrP undergoes a profound structural transition during prion propagation, it seems likely that other proteins such as chaperones participate in this process. Whether protein X functions as a molecular chaperone is unknown. Interestingly, scrapie-infected cells in culture display marked differences in the induction of heat-shock proteins (234, 235), and Hsp70 mRNA has been reported to increase in scrapie of mice (236). While attempts to isolate specific proteins that bind to PrP have been disappointing (237), PrP has been shown to interact with Bcl-2 and Hsp60 by two-hybrid analysis in yeast (238, 239). Although these studies are suggestive, no molecular chaperone involved in prion formation in mammalian cells has been identified.
Miniprions.
By using the four-helix bundle model of PrPC (Fig. (Fig.44A) (147), each region of proposed secondary structure was systematically deleted and the mutant constructs were expressed in scrapie-infected neuroblastoma (ScN2a) cells and Tg mice (164, 168). Deletion of any of the four putative helical regions prevented PrPSc formation, whereas deletion of the NH2-terminal region containing residues 23–89 did not affect the yield of PrPSc. In addition to the 67 residues at the NH2 terminus, 36 residues from positions 141–176 could be deleted without altering PrPSc formation (Figs. (Figs.66 and and7).7). The resulting PrP molecule of 106 amino acids was designated PrP106. In this mutant PrP, helix A as well as the S2 β-strand were removed. When PrP106 was expressed in ScN2a cells, PrPSc106 was soluble in 1% Sarkosyl. Whether the structure of PrPSc106 can be more readily determined than that of full-length PrPSc remains uncertain.
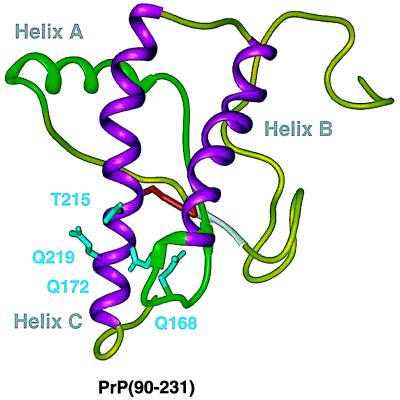
Miniprions produced by deleting PrP residues 23–89 and 141–176. The deletion of residues 141–176 (green) containing helix A and the S2 β-strand is shown. Side chains of residues 168, 172, 215, and 219, which are thought to bind protein X, are shown in cyan.
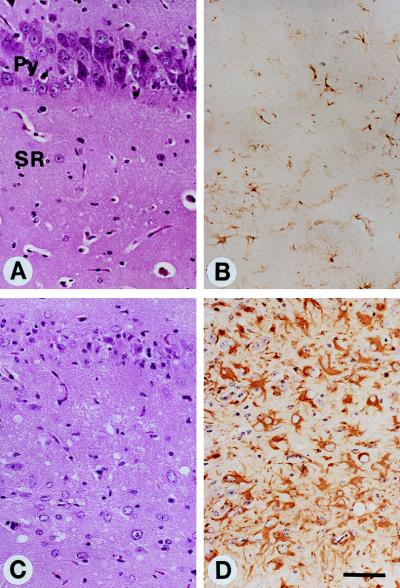
Tg(PrP106)Prnp0/0 mice were inoculated with RML106 prions containing PrPSc106. Sections of the hippocampus were stained with hematoxylin and eosin (A and C) and immunostained for GFAP (B and D). (A and B) Control Tg(PrP106)Prnp0/0 mouse uninoculated and without neurologic deficits. (C and D) Tg(PrP106)Prnp0/0 mouse inoculated with RML106 prions and sacrificed after signs of neurologic dysfunction were observed. (Bar = 50 μm.) Photomicrographs prepared by Stephen J. DeArmond.
Transgene-specified susceptibility.
Tg(PrP106)Prnp0/0 mice that expressed PrP106 developed neurological dysfunction ≈300 days after inoculation with RML prions previously passaged in CD-1 Swiss mice (S. Supattapone, T. Muramoto, D. Peretz, S. J. DeArmond, A. Wallace, F. E. Cohen, S.B.P., and M. R. Scott, unpublished results). The resulting prions containing PrPSc106 produced CNS disease in ≈66 days upon subsequent passage in Tg(PrP106)Prnp0/0 mice (Table (Table5).5). Besides widespread spongiform degeneration and PrP deposits, the pyramidal cells of the hippocampus constituting the CA-1, CA-2, and CA-3 fields disappeared in Tg(PrP106)Prnp0/0 mice inoculated with prions containing PrPSc106 (Fig. (Fig.7).7). In no previous study of Tg mice have we seen similar neuropathological lesions. The Tg(MoPrP-A) mice overexpressing MoPrP are resistant to RML106 miniprions but are highly susceptible to RML prions. These mice require more than 250 days to produce illness after inoculation with miniprions but develop disease in ≈50 days when inoculated with RML prions containing full-length MoPrPSc.
Table 5
Susceptibility and resistance of Tg mice to
artificial miniprions
miniprions
| Host | Incubation time, days
(n/n0)
| |
|---|---|---|
| RML106 miniprions | RML prions | |
| Tg(PrP106)Prnp0/0 mice | 66 ± ± 3 3 (10/10) (10/10) | 300 ±
± 2 2 (9/10) (9/10) |
| Tg(MoPrP-A) mice | >250 (0/11) (0/11) | 50 ± ± 2
2 (16/16) (16/16) |
Data are from ref. 218 and S. Supattapone, T. Muramoto, D. Peretz, S. J. DeArmond, A. Wallace, F. E. Cohen, S.B.P., and M. R. Scott, unpublished work. Incubation times are given ±SEM.
Smaller prions and mythical viruses.
The unique incubation times and neuropathology in Tg mice caused by miniprions are difficult to reconcile with the notion that scrapie is caused by an as-yet-unidentified virus. When the mutant or wt PrPC of the host matched PrPSc in the inoculum, the mice were highly susceptible (Table (Table5).5). However, when there was a mismatch between PrPC and PrPSc, the mice were resistant to the prions. This principle of homologous PrP interactions, which underlies the species barrier (Table (Table3),3), is recapitulated in studies of PrP106 where the amino acid sequence has been drastically changed by deleting nearly 50% of the residues. Indeed, the unique properties of the miniprions provide another persuasive argument supporting the contention that prions are infectious proteins.
Human Prion Diseases.
Most humans afflicted with prion disease present with rapidly progressive dementia, but some manifest cerebellar ataxia. Although the brains of patients appear grossly normal upon postmortem examination, they usually show spongiform degeneration and astrocytic gliosis under the microscope. The human prion diseases can present as sporadic, genetic, or infectious disorders (5) (Table (Table11).
Sporadic CJD.
Sporadic forms of prion disease constitute most cases of CJD and possibly a few cases of Gerstmann–Sträussler–Scheinker disease (GSS) (Table (Table1)1) (4, 240, 241). In these patients, mutations of the PrP gene are not found. How prions causing disease arise in patients with sporadic forms is unknown; hypotheses include horizontal transmission of prions from humans or animals (242), somatic mutation of the PrP gene, and spontaneous conversion of PrPC into PrPSc (5, 15). Because numerous attempts to establish an infectious link between sporadic CJD and a preexisting prion disease in animals or humans have been unrewarding, it seems unlikely that transmission features in the pathogenesis of sporadic prion disease (9–12, 243).
Inherited prion diseases.
To date, 20 different mutations in the human PrP gene resulting in nonconservative substitutions have been found that segregate with the inherited prion diseases (Fig. (Fig.44B). Familial CJD cases suggested that genetic factors might influence pathogenesis (1, 2, 244), but this was difficult to reconcile with the transmissibility of fCJD and GSS (3). The discovery of genetic linkage between the PrP gene and scrapie incubation times in mice (213) raised the possibility that mutation might feature in the hereditary human prion diseases. The P102L mutation was the first PrP mutation to be genetically linked to CNS dysfunction in GSS (Fig. (Fig.44B) (4) and has since been found in many GSS families throughout the world (245–247). Indeed, a mutation in the protein coding region of the PrP gene has been found in all reported kindred with familial human prion disease; besides the P102L mutation, genetic linkage has been established for four other mutations (16, 29–31).
Tg mouse studies confirmed that mutations of the PrP gene can cause neurodegeneration. The P102L mutation of GSS was introduced into the MoPrP transgene, and five lines of Tg(MoPrP-P101L) mice expressing high levels of mutant PrP developed spontaneous CNS degeneration consisting of widespread vacuolation of the neuropil, astrocytic gliosis, and numerous PrP amyloid plaques similar to those seen in the brains of humans who die from GSS(P102L) (248–250). Brain extracts prepared from spontaneously ill Tg(MoPrP-P101L) mice transmitted CNS degeneration to Tg196 mice but contained no protease-resistant PrP (249, 250). The Tg196 mice do not develop spontaneous disease but express low levels of the mutant transgene MoPrP-P101L and are deficient for mouse PrP (Prnp0/0) (221). These studies, combined with the transmission of prions from patients who died of GSS to apes and monkeys (3) or to Tg(MHu2M-P101L) mice (134), demonstrate that prions are generated de novo by mutations in PrP. Additionally, brain extracts from patients with some other inherited prion diseases, fCJD(E200K) or FFI, transmit disease to Tg(MHu2M) mice (27). An artificial set of mutations in a PrP transgene consisting of A113V, A115V, and A118V produced neurodegeneration in neonatal mice; these Val substitutions were selected for their propensity to form β-sheets (153, 251). In preliminary studies, brain extracts from two of these mice transmitted disease to hamsters and to Tg mice expressing a chimeric SHa/Mo PrP.
Genetic disease that is transmissible.
Had the PrP gene been identified in families with prion disease by positional cloning or through the purification and sequencing of PrP in amyloid plaques before brain extracts were shown to be transmissible, the prion concept might have been more readily accepted. Within that scenario, it seems likely that we would have explored the possibility that the mutant protein, upon inoculation into a susceptible host, stimulated production of more of a similar protein. Postulating an infectious pathogen with a foreign genome would have been the least likely candidate to explain how a genetic disease could be experimentally transmissible.
Infectious prion diseases.
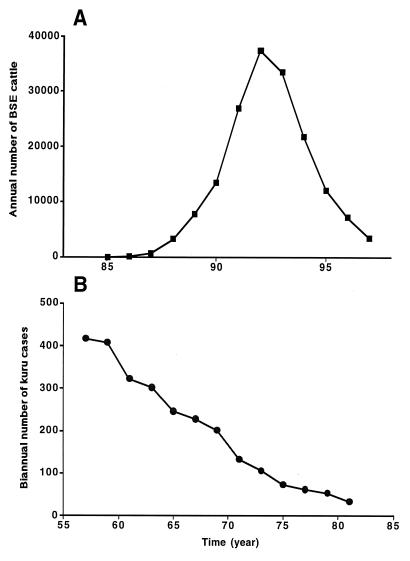
The infectious prion diseases include kuru of the Fore people in New Guinea, where prions were transmitted by ritualistic cannibalism (242, 252, 253). With the cessation of cannibalism at the urging of missionaries, kuru began to decline long before it was known to be transmissible (Fig. (Fig.8).8). Sources of prions causing infectious CJD on several different continents include improperly sterilized depth electrodes, transplanted corneas, human growth hormone (HGH) and gonadotropin derived from cadaveric pituitaries, and dura mater grafts (254). Over 90 young adults have developed CJD after treatment with cadaveric HGH; the incubation periods range from 3 to more than 20 years (255, 256). Dura mater grafts implanted during neurosurgical procedures seem to have caused more than 60 cases of CJD; these incubation periods range from 1 to more than 14 years (257–259).
Prion Diversity.
The existence of prion strains raises the question of how heritable biological information can be enciphered in a molecule other than nucleic acid (131, 215, 260–264). Strains or varieties of prions have been defined by incubation times and the distribution of neuronal vacuolation (215, 265). Subsequently, the patterns of PrPSc deposition were found to correlate with vacuolation profiles, and these patterns were also used to characterize strains of prions (231, 266, 267).
The typing of prion strains in C57BL, VM, and (C57BL × VM)F1 inbred mice began with isolates from sheep with scrapie. The prototypic strains called Me7 and 22A gave incubation times of ≈150 and ≈400 days in C57BL mice, respectively (215, 268, 269). The PrP genes of C57BL and I/Ln (and later VM) mice encode proteins differing at two residues and control scrapie incubation times (195, 213, 217–219, 270).
Until recently, support for the hypothesis that the tertiary structure of PrPSc enciphers strain-specific information (23) was minimal except for the DY strain isolated from mink with transmissible encephalopathy by passage in Syrian hamsters (26, 271, 272). PrPSc in DY prions showed diminished resistance to proteinase K digestion as well as a peculiar site of cleavage. The DY strain presented a puzzling anomaly, since other prion strains exhibiting similar incubation times did not show this altered susceptibility to proteinase K digestion of PrPSc (135). Also notable was the generation of new strains during passage of prions through animals with different PrP genes (135, 230).
PrPSc conformation enciphers diversity.
Persuasive evidence that strain-specific information is enciphered in the tertiary structure of PrPSc comes from transmission of two different inherited human prion diseases to mice expressing a chimeric MHu2M PrP transgene (27). In FFI, the protease-resistant fragment of PrPSc after deglycosylation has a mass of 19 kDa, whereas in fCJD(E200K) and most sporadic prion diseases it is 21 kDa (Table (Table6)6) (273, 274). This difference in molecular size was shown to be due to different sites of proteolytic cleavage at the NH2 termini of the two human PrPSc molecules reflecting different tertiary structures (273). These distinct conformations were understandable because the amino acid sequences of the PrPs differ.
Table 6
Distinct prion strains generated in humans with inherited
prion diseases and transmitted to Tg mice
mice
| Inoculum | Host species | Host PrP genotype | Incubation time, days | (n/n0) | PrPSc, kDa |
|---|---|---|---|---|---|
| None | Human | FFI(D178N, M129) | 19 | ||
| FFI | Mouse | Tg(MHu2M) | 206 ± ± 7
7 |  (7/7) (7/7) | 19 |
| FFI → Tg(MHu2M) | Mouse | Tg(MHu2M) | 136 ± ± 1 1 |  (6/6) (6/6) | 19 |
| None | Human | fCJD(E200K) | 21 | ||
| fCJD | Mouse | Tg(MHu2M) | 170 ± ± 2 2 | (10/10) | 21 |
| fCJD → Tg(MHu2M) | Mouse | Tg(MHu2M) | 167 ± ± 3
3 | (15/15) | 21 |
Data are from ref. 27 and G. Telling and S.B.P., unpublished work. Incubation times are given ±SEM.
Extracts from the brains of FFI patients transmitted disease to mice expressing a chimeric MHu2M PrP gene about 200 days after inoculation and induced formation of the 19-kDa PrPSc, whereas fCJD(E200K) and sCJD produced the 21-kDa PrPSc in mice expressing the same transgene (27). On second passage, Tg(MHu2M) mice inoculated with FFI prions showed an incubation time of ≈130 days and a 19-kDa PrPSc, whereas those inoculated with fCJD(E200K) prions exhibited an incubation time of ≈170 days and a 21-kDa PrPSc (28). The experimental data demonstrate that MHu2MPrPSc can exist in two different conformations based on the sizes of the protease-resistant fragments, yet the amino acid sequence of MHu2MPrPSc is invariant.
The results of our studies argue that PrPSc acts as a template for the conversion of PrPC into nascent PrPSc. Imparting the size of the protease-resistant fragment of PrPSc through conformational templating provides a mechanism for both the generation and propagation of prion strains.
Interestingly, the protease-resistant fragment of PrPSc after deglycosylation with a mass of 19 kDa has been found in a patient who developed a sporadic case of prion disease similar to FFI but with no family history. Because both PrP alleles encoded the wt sequence and a Met at position 129, we labeled this case fatal sporadic insomnia (FSI). At autopsy, the spongiform degeneration, reactive astrogliosis, and PrPSc deposition were confined to the thalamus (275). These findings argue that the clinicopathologic phenotype is determined by the conformation of PrPSc in accord with the results of the transmission of human prions from patients with FFI to Tg mice (27).
Selective neuronal targeting.
Besides incubation times, profiles of spongiform change (Fig. (Fig.1)1) have been used to characterize prion strains (276), but recent studies argue that such profiles are not an intrinsic feature of strains (277, 278). The mechanism by which prion strains modify the pattern of spongiform degeneration was perplexing, since earlier investigations had shown that PrPSc deposition precedes neuronal vacuolation and reactive gliosis (212, 231). When FFI prions were inoculated into Tg(MHu2M) mice, PrPSc was confined largely to the thalamus (Fig. (Fig.99A) as is the case for FFI in humans (27, 279). In contrast, fCJD(E200K) prions inoculated into Tg(MHu2M) mice produced widespread deposition of PrPSc throughout the cortical mantle and many of the deep structures of the CNS (Fig. (Fig.99B) as is seen in fCJD(E200K) of humans. To examine whether the diverse patterns of PrPSc deposition are influenced by Asn-linked glycosylation of PrPC, we constructed Tg mice expressing PrPs mutated at one or both of the Asn-linked glycosylation consensus sites (278). These mutations resulted in aberrant neuroanatomic topologies of PrPC within the CNS, whereas pathologic point mutations adjacent to the consensus sites did not alter the distribution of PrPC. Tg mice with mutation of the second PrP glycosylation site exhibited prion incubation times of >500 days and unusual patterns of PrPSc deposition. These findings raise two possible scenarios. First, glycosylation can modify the conformation of PrPC and affect its affinity for a particular conformer of PrPSc, which results in specific patterns of PrPSc deposition; such interactions between PrPSc and PrPC are likely to determine the rate of nascent PrPSc formation. Second, glycosylation modifies the stability of PrPSc and, hence, the rate of PrPSc clearance. This latter explanation is consistent with the proposal that the binding of PrPC to protein X is the rate-limiting step in PrPSc formation under most circumstances (280, 345).
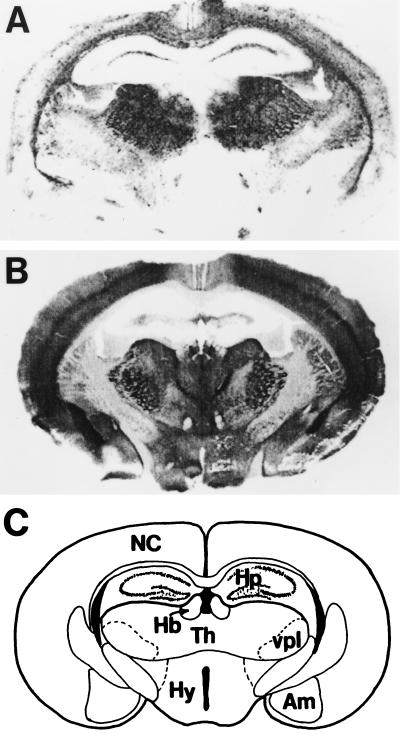
Regional distribution of PrPSc deposition in Tg(MHu2M)Prnp0/0 mice inoculated with prions from humans who died of inherited prion diseases (Table (Table5).5). Histoblot of PrPSc deposition in a coronal section a Tg(MHu2M)Prnp0/0 mouse through the hippocampus and thalamus (27). (A) The Tg mouse was inoculated with brain extract prepared from a patient who died of FFI. (B) The Tg mouse was inoculated with extract from a patient with fCJD(E200K). Cryostat sections were mounted on nitrocellulose and treated with proteinase K to eliminate PrPC (209). To enhance the antigenicity of PrPSc, the histoblots were exposed to 3 M guanidinium isothiocyanate before immunostaining using anti-PrP 3F4 mAb (174). (C) Labeled diagram of a coronal sections of the hippocampus/thalamus region. NC, neocortex; Hp, hippocampus; Hb, habenula; Th, thalamus; vpl, ventral posterior lateral thalamic nucleus; Hy, hypothalamus; Am, amygdala. Photomicrographs were prepared by Stephen J. DeArmond.
BSE.
Prion strains and the species barrier are of paramount importance in understanding the BSE epidemic in Great Britain, in which it is estimated that almost one million cattle were infected with prions (281, 282). The mean incubation time for BSE is about 5 years. Most cattle therefore did not manifest disease because they were slaughtered between 2 and 3 years of age (283). Nevertheless, more than 160,000 cattle, primarily dairy cows, have died of BSE over the past decade (Fig. (Fig.88A) (281). BSE is a massive common-source epidemic caused by meat and bone meal (MBM) fed primarily to dairy cows (282, 284). The MBM was prepared from the offal of sheep, cattle, pigs, and chickens as a high-protein nutritional supplement. In the late 1970s, the hydrocarbon-solvent extraction method used in the rendering of offal began to be abandoned, resulting in MBM with a much higher fat content (284). It is now thought that this change in the rendering process allowed scrapie prions from sheep to survive rendering and to be passed into cattle. Alternatively, bovine prions were present at low levels prior to modification of the rendering process and with the processing change survived in sufficient numbers to initiate the BSE epidemic when inoculated back into cattle orally through MBM. Against the latter hypothesis is the widespread geographical distribution throughout England of the initial 17 cases of BSE, which occurred almost simultaneously (282, 285, 286). Furthermore, there is no evidence of a preexisting prion disease of cattle, either in Great Britain or elsewhere.
Origin of BSE prions?
The origin of the bovine prions causing BSE cannot be determined by examining the amino acid sequence of PrPSc in cattle with BSE because the PrPSc in these animals has the bovine sequence whether the initial prions in MBM came from cattle or sheep. The bovine PrP sequence differs from that of sheep at 7 or 8 positions (287–289). In contrast to the many PrP polymorphisms found in sheep, only one PrP polymorphism has been found in cattle. Though most bovine PrP alleles encode five octarepeats, some encode six. PrP alleles encoding six octarepeats do not seem to be overrepresented in BSE (Fig. (Fig.44B) (290).
Brain extracts from BSE cattle cause disease in cattle, sheep, mice, pigs, and mink after intracerebral inoculation (291–295), but prions in brain extracts from sheep with scrapie fed to cattle produced illness substantially different from BSE (296). However, no exhaustive effort has been made to test different strains of sheep prions or to examine the disease after bovine-to-bovine passage. The annual incidence of sheep with scrapie in Britain over the past two decades has remained relatively low (J. Wilesmith, personal communication). In July 1988, the practice of feeding MBM to sheep and cattle was banned. Recent statistics argue that the epidemic is now disappearing as a result of this ruminant feed ban (Fig. (Fig.88A) (281), reminiscent of the disappearance of kuru in the Fore people of New Guinea (242, 253) (Fig. (Fig.88B).
Monitoring cattle for BSE prions.
Although many plans have been offered for the culling of older cattle to minimize the spread of BSE (281), it seems more important to monitor the frequency of prion disease in cattle as they are slaughtered for human consumption. No reliable, specific test for prion disease in live animals is available, but immunoassays for PrPSc in the brainstems of cattle might provide a reasonable approach to establishing the incidence of subclinical BSE in cattle entering the human food chain (176, 209, 289, 297–299, 345). Determining how early in the incubation period PrPSc can be detected by immunological methods is now possible, since a reliable bioassay has been created by expressing the BoPrP gene in Tg mice (180). Prior to development of Tg(BoPrP)Prnp0/0 mice, non-Tg mice inoculated intracerebrally with BSE brain extracts required more than 300 days to develop disease (67, 295, 300, 301). Depending on the titer of the inoculum, the structures of PrPC and PrPSc, and the structure of protein X, the number of inoculated animals developing disease can vary over a wide range (Table (Table3).3). Some investigators have stated that transmission of BSE to mice is quite variable with incubation periods exceeding 1 year (67), whereas others report low prion titers in BSE brain homogenates (300, 301) compared with rodent brain scrapie (54, 56, 302, 303).
Have bovine prions been transmitted to humans?
In 1994, the first cases of CJD in teenagers and young adults that were eventually labeled new variant (v) CJD occurred in Britain (304). Besides these patients being young (305, 306), their brains showed numerous PrP amyloid plaques surrounded by a halo of intense spongiform degeneration (Fig. (Fig.10)10) (307). Later, one French case meeting these criteria followed (308). These unusual neuropathologic changes have not been seen in CJD cases in the United States, Australia, or Japan (307, 309). Both macaque monkeys and marmosets developed neurologic disease several years after inoculation with bovine prions (310), but only the macaques exhibited numerous PrP plaques similar to those found in vCJD (311) (R. Ridley and H. Baker, personal communication).
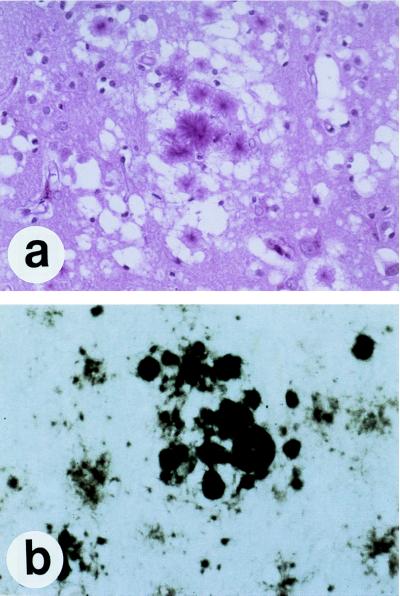
Histopathology of vCJD in Great Britain. (a) Section from frontal cortex stained by the periodic acid–Schiff method, showing a field with aggregates of plaques surrounded by spongiform degeneration. (×93.) (b) Multiple plaques and amorphous deposits are PrP immunopositive. (×460.) Specimens were provided by James Ironside, Jeanne Bell, and Robert Will; photomicrographs were prepared by Stephen J. DeArmond.
The restricted geographical occurrence and chronology of vCJD have raised the possibility that BSE prions have been transmitted to humans. That only ≈25 vCJD cases have been recorded and the incidence has remained relatively constant make establishing the origin of vCJD difficult. No set of dietary habits distinguishes vCJD patients from apparently healthy people. Moreover, there is no explanation for the predilection of vCJD for teenagers and young adults. Why have older individuals not developed vCJD-based neuropathologic criteria? It is noteworthy that epidemiological studies over the past three decades have failed to find evidence for transmission of sheep prions to humans (9–12). Attempts to predict the future number of cases of vCJD, assuming exposure to bovine prions prior to the offal ban, have been uninformative because so few cases of vCJD have occurred (312–314). Are we at the beginning of a human prion disease epidemic in Britain like those seen for BSE and kuru (Fig. (Fig.8),8), or will the number of vCJD cases remain small as seen with iCJD caused by cadaveric HGH (255, 256)?
Strain of BSE prions.
Was a particular conformation of bovine PrPSc selected for heat resistance during the rendering process and then reselected multiple times as cattle infected by ingesting prion-contaminated MBM were slaughtered and their offal rendered into more MBM? Recent studies of PrPSc from brains of patients who died of vCJD show a pattern of PrP glycoforms different from those found for sCJD or iCJD (315, 316). But the utility of measuring PrP glycoforms is questionable in trying to relate BSE to vCJD (317, 318) because PrPSc is formed after the protein is glycosylated (138, 140) and enzymatic deglycosylation of PrPSc requires denaturation (319, 320). Alternatively, it may be possible to establish a relationship between the conformations of PrPSc from cattle with BSE and those from humans with vCJD by using Tg mice as was done for strains generated in the brains of patients with FFI or fCJD (27, 180). A relationship between vCJD and BSE has been suggested by finding similar incubation times in non-Tg RIII mice of ≈310 days after inoculation with Hu or Bo prions (295).
Yeast and Other Prions.
Although prions were originally defined in the context of an infectious pathogen (58), it is now becoming widely accepted that prions are elements that impart and propagate variability through multiple conformers of a normal cellular protein. Such a mechanism must surely not be restricted to a single class of transmissible pathogens. Indeed, it is likely that the original definition will need to be extended to encompass other situations in which a similar mechanism of information transfer occurs.
Two notable prion-like determinants, [URE3] and [PSI], have already been described in yeast and one in another fungus denoted [Het-s*] (321–326). Studies of candidate prion proteins in yeast may prove particularly helpful in the dissection of some of the events that feature in PrPSc formation. Interestingly, different strains of yeast prions have been identified (327). Conversion to the prion-like [PSI] state in yeast requires the molecular chaperone Hsp104; however, no homolog of Hsp104 has been found in mammals (322, 328). The NH2-terminal prion domains of Ure2p and Sup35 that are responsible for the [URE3] and [PSI] phenotypes in yeast have been identified. In contrast to PrP, which is a GPI-anchored membrane protein, both Ure2p and Sup35 are cytosolic proteins (329). When the prion domains of these yeast proteins were expressed in E. coli, the proteins were found to polymerize into fibrils with properties similar to those of PrP and other amyloids (323–325).
Whether prions explain some other examples of acquired inheritance in lower organisms is unclear (330, 331). For example, studies on the inheritance of positional order and cellular handedness on the surface of small organisms have demonstrated the epigenetic nature of these phenomena, but the mechanism remains unclear (332, 333).
Prevention of and Therapeutics for Prion Diseases.
As our understanding of prion propagation increases, it should be possible to design effective therapeutics. Because people at risk for inherited prion diseases can now be identified decades before neurologic dysfunction is evident, the development of an effective therapy for these fully penetrant disorders is imperative (334, 335). Although we have no way of predicting the number of individuals who may develop neurologic dysfunction from bovine prions in the future (313), it would be prudent to seek an effective therapy now (28, 336). Interfering with the conversion of PrPC into PrPSc seems to be the most attractive therapeutic target (60). Either stabilizing the structure of PrPC by binding a drug or modifying the action of protein X, which might function as a molecular chaperone (Fig. (Fig.55B), is a reasonable strategy. Whether it is more efficacious to design a drug that binds to PrPC at the protein X binding site or one that mimics the structure of PrPC with basic polymorphic residues that seem to prevent scrapie and CJD remains to be determined (178, 233). Because PrPSc formation seems limited to caveolae-like domains (142–145, 337), drugs designed to inhibit this process need not penetrate the cytosol of cells, but they do need to be able to enter the CNS. Alternatively, drugs that destabilize the structure of PrPSc might also prove useful.
The production of domestic animals that do not replicate prions may also be important with respect to preventing prion disease. Sheep encoding the R/R polymorphism at position 171 seem to be resistant to scrapie (199, 232, 338–344); presumably, this was the genetic basis of Parry’s scrapie eradication program in Great Britain 30 years ago (44, 46). A more effective approach using dominant negatives for producing prion-resistant domestic animals, including sheep and cattle, is probably the expression of PrP transgenes encoding R171 as well as additional basic residues at the putative protein X binding site (Fig. (Fig.55B) (178). Such an approach can be readily evaluated in Tg mice, and once shown to be effective, it could be instituted by artificial insemination of sperm from males homozygous for the transgene. More difficult is the production of PrP-deficient cattle and sheep. Although such animals would not be susceptible to prion disease (100, 101), they might suffer some deleterious effects from ablation of the PrP gene (223–225, 228).
Whether gene therapy for the human prion diseases by using the dominant-negative approach described above for prion-resistant animals will prove feasible depends on the availability of efficient vectors for delivery of the transgene to the CNS.
Concluding Remarks—Looking to the Future.
Although the study of prions has taken several unexpected directions over the past three decades, a rather novel and fascinating story of prion biology is emerging. Investigations of prions have elucidated a previously unknown mechanism of disease in humans and animals. While learning the details of the structures of PrPs and deciphering the mechanism of PrPC transformation into PrPSc will be important, the fundamental principles of prion biology have become reasonably clear. Though some investigators prefer to view the composition of the infectious prion particle as unresolved (336, 346), such a perspective denies an enlarging body of data, none of which refute the prion concept. Moreover, the discovery of prion-like phenomena mediated by proteins unrelated to PrP in yeast and fungi serves not only to strengthen the prion concept but also to widen it (329).
Hallmark of prion diseases.
The hallmark of all prion diseases—whether sporadic, dominantly inherited, or acquired by infection—is that they involve the aberrant metabolism and resulting accumulation of the prion protein (Table (Table1)1) (23). The conversion of PrPC into PrPSc involves a conformation change whereby the α-helical content diminishes and the amount of β-sheet increases (25). These findings provide a reasonable mechanism to explain the conundrum presented by the three different manifestations of prion disease.
Understanding how PrPC unfolds and refolds into PrPSc will be of paramount importance in transferring advances in the prion diseases to studies of other degenerative illnesses. The mechanism by which PrPSc is formed must involve a templating process whereby existing PrPSc directs the refolding of PrPC into a nascent PrPSc with the same conformation. A knowledge of PrPSc formation not only will help in the rational design of drugs that interrupt the pathogenesis of prion diseases but it may also open new approaches to deciphering the causes of and to developing effective therapies for the more common neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (ALS). Indeed, the expanding list of prion diseases and their novel modes of transmission and pathogenesis (Table (Table1),1), as well as the unprecedented mechanisms of prion propagation and information transfer (Tables (Tables55 and and6),6), indicate that much more attention to these fatal disorders of protein conformation is urgently needed.
But prions may have even wider implications than those noted for the common neurodegenerative diseases. If we think of prion diseases as disorders of protein conformation and do not require the diseases to be transmissible, then what we have learned from the study of prions may reach far beyond these common illnesses.
Multiple conformers.
The discovery that proteins may have multiple biologically active conformations may prove no less important than the implications of prions for diseases. How many different tertiary structures can PrPSc adopt? This query not only addresses the issue of the limits of prion diversity (345) but also applies to proteins as they normally function within the cell or act to affect homeostasis in multicellular organisms. The expanding list of chaperones that assist the folding and unfolding of proteins promises much new knowledge about this process. For example, it is now clear that proproteases can carry their own chaperone activity where the pro portion of the protein functions as a chaperone in cis to guide the folding of the proteolytically active portion before it is cleaved (347). Such a mechanism might well feature in the maturation of polypeptide hormones. Interestingly, mutation of the chaperone portion of prosubtilisin resulted in the folding of a subtilisin protease with different properties than the one folded by the wild-type chaperone. Such chaperones have also been shown to work in trans (347). Besides transient metabolic regulation within the cell and hormonal regulation of multicellular organisms, it is not unreasonable to suggest that polymerization of proteins into multimeric structures such as intermediate filaments might be controlled at least in part by alternative conformations of proteins. Such regulation of multimeric protein assemblies might occur either in the proteins that form the polymers or in the proteins that function to facilitate the polymerization process. Additionally, apoptosis during development and throughout adult life might also be regulated, at least in part, by alternative tertiary structures of proteins.
Shifting the debate.
The debate about prions and the diseases that they cause has now shifted to such issues as how many biological processes are controlled by changes in protein conformation. Although the extreme radiation-resistance of the scrapie infectivity suggested that the pathogen causing this disease and related illnesses would be different from viruses, viroids, and bacteria (32, 33), few thought that alternative protein conformations might even remotely feature in the pathogenesis of the prion diseases (37). Indeed, an unprecedented mechanism of disease has been revealed in which an aberrant conformational change in a protein is propagated. The discovery of prions and their eventual acceptance by the community of scholars represents a triumph of the scientific process over prejudice. The future of this new and evolving area of biology should prove even more interesting and productive as a multitude of unpredicted discoveries emerge.
Acknowledgments
I thank O. Abramsky, M. Baldwin, R. Barry, C. Bellinger, D. Borchelt, D. Burton, D. Butler, G. Carlson, J. Cleaver, F. Cohen, C. Cooper, S. DeArmond, T. Diener, J. Dyson, S. Farr-Jones, R. Fletterick, R. Gabizon, D. Groth, C. Heinrich, L. Hood, K. Hsiao, Z. Huang, T. James, K. Kaneko, R. Kohler, T. Kitamoto, H. Kretzschmar, V. Lingappa, H. Liu, D. Lowenstein, M. McKinley, Z. Meiner, B. Miller, W. Mobley, B. Oesch, K.-M. Pan, D. Peretz, D. Riesner, G. Roberts, J. Safar, M. Scott, A. Serban, N. Stahl, A. Taraboulos, J. Tateishi, M. Torchia, P. Tremblay, M. Vey, C. Weissmann, D. Westaway, A. Williamson, P. Wright, and L. Zullianello for their contributions to many phases of these studies and H. Baron, W. Burke, I. Diamond, H. Fields, R. Fishman, D. Gilden, S. Hauser, H. Koprowski, J. Krevans, J. Martin, M. McCarty, R. C. Morris, N. Nathanson, F. Seitz, and C. Yanofsky for support and encouragement. This research was supported by grants from the National Institute of Aging and the National Institute of Neurologic Diseases and Stroke of the National Institutes of Health, the National Science Foundation, International Human Frontiers of Science Program, and the American Health Assistance Foundation, as well as by gifts from the Sherman Fairchild Foundation, the Keck Foundation, the G. Harold and Leila Y. Mathers Foundation, the Bernard Osher Foundation, the John D. French Foundation, the Howard Hughes Medical Institute, R. J. Reynolds, National Medical Enterprises, and Centeon.
ABBREVIATIONS
| Bo | bovine |
| BSE | bovine spongiform encephalopathy |
| CJD | Creutzfeldt–Jakob disease |
| sCJD | sporadic CJD |
| fCJD | familial CJD |
| iCJD | iatrogenic CJD |
| vCJD | (new) variant CJD |
| CNS | central nervous system |
| CWD | chronic wasting disease |
| FFI | fatal familial insomnia |
| FSE | feline spongiform encephalopathy |
| FSI | fatal sporadic insomnia |
| GFAP | glial fibrillary acidic protein |
| GPI | glycosyl-phosphatidylinositol |
| GSS | Gerstmann–Sträussler–Scheinker disease |
| HGH | human growth hormone |
| Hu | human |
| MBM | meat and bone meal |
| Mo | mouse |
| r | recombinant |
| SHa | Syrian hamster |
| Tg | transgenic |
| TME | transmissible mink encephalopathy |
| wt | wild-type |
Footnotes
Adapted from Les Prix Nobel, 1997. © 1997 by The Nobel Foundation
Editor’s Note: This article is an abbreviated version of Stanley B. Prusiner’s Nobel Lecture, “Prions.” The 1997 Nobel Prize in Physiology or Medicine was awarded to Dr. Prusiner for his discovery of prions, an entirely new genre of disease-causing agents, and for elucidating the fascinating principles that underline their mode of action. The Nobel Foundation graciously has granted us permission to reprint this article. The Nobel Lectures provide examples of successful approaches to major scientific problems as well as authoritative reviews. However, in recent years, these lectures have rarely been read, perhaps because of the difficulty in obtaining the collections. By reprinting this lecture we hope to broaden their exposure.
‡Prions are defined as proteinaceous infectious particles that lack nucleic acid. PrPC is the cellular prion protein; PrPSc is the pathologic isoform. NH2-terminal truncation during limited proteolysis of PrPSc produces PrP 27–30.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.95.23.13363
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC33918
Citations & impact
Impact metrics
Article citations
Copper binding alters the core structure of amyloid fibrils formed by Y145Stop human prion protein.
Phys Chem Chem Phys, 26(41):26489-26496, 23 Oct 2024
Cited by: 0 articles | PMID: 39392708 | PMCID: PMC11469299
Viroids and Satellites and Their Vector Interactions.
Viruses, 16(10):1598, 11 Oct 2024
Cited by: 0 articles | PMID: 39459931 | PMCID: PMC11512221
Potential of Marine Sponge Metabolites against Prions: Bromotyrosine Derivatives, a Family of Interest.
Mar Drugs, 22(10):456, 04 Oct 2024
Cited by: 0 articles | PMID: 39452864 | PMCID: PMC11509309
Abnormal synaptic architecture in iPSC-derived neurons from a multi-generational family with genetic Creutzfeldt-Jakob disease.
Stem Cell Reports, 19(10):1474-1488, 26 Sep 2024
Cited by: 0 articles | PMID: 39332406 | PMCID: PMC11561462
Prion-like Proteins in Plants: Key Regulators of Development and Environmental Adaptation via Phase Separation.
Plants (Basel), 13(18):2666, 23 Sep 2024
Cited by: 0 articles | PMID: 39339640 | PMCID: PMC11435361
Review Free full text in Europe PMC
Go to all (3,527) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetics of prions.
Annu Rev Genet, 31:139-175, 01 Jan 1997
Cited by: 72 articles | PMID: 9442893
Review
Genetic and infectious prion diseases.
Arch Neurol, 50(11):1129-1153, 01 Nov 1993
Cited by: 87 articles | PMID: 8105771
Review
Prion encephalopathies of animals and humans.
Dev Biol Stand, 80:31-44, 01 Jan 1993
Cited by: 13 articles | PMID: 8270114
Review
Prion diseases and the BSE crisis.
Science, 278(5336):245-251, 01 Oct 1997
Cited by: 451 articles | PMID: 9323196
Review