Abstract
Free full text

Evidence That γδ versus αβ T Cell Fate Determination Is Initiated Independently of T Cell Receptor Signaling
Abstract
Two types of T cells, αβ and γδ, develop in vertebrates. How these two T cell lineages arise from a common thymic T progenitor is poorly understood. Differentiation of αβ lineage T cells requires the surrogate α chain (pTα), which associates with the T cell receptor (TCR) β chain to form the pre-TCR. γδ lineage development does not appear to involve an obligatory surrogate chain, but instead requires productive rearrangement and expression of both TCR γ and δ genes. It has been proposed that the quality of signals transmitted by the pre-TCR and γδ TCR are distinct and that these “instructive” signals determine the lineage fate of an uncommitted progenitor cell. Here we show that the thymic T progenitor cells (CD25+CD44+c-kit+CD3−CD4−CD8− thymocytes, termed pro-T cells) from young adult mice that have yet to express TCRs can be subdivided based on interleukin 7 receptor (IL-7R) expression. These subsets exhibit differential potential to develop into γδ versus αβ lineage (CD4+CD8+ cells) in the thymus. Upon intrathymic injection, IL-7Rneg-lo pro-T cells generated a 13-fold higher ratio of αβ lineage to γδ lineage cells than did IL-7R+ pro-T cells. Much of this difference was due to a fivefold greater potential of IL-7R+ pro-T cells to develop into TCR-γδ T cells. Evidence indicates that this biased developmental potential is not a result of enhanced TCR-γ gene rearrangement/expression in IL-7R+ pro-T cells. These results indicate that the pro-T cells are heterogeneous in developmental potential before TCR gene rearrangement and suggest that in some precursor cells the initial lineage commitment is independent of TCR-mediated signals.
Introduction
The mechanism of T cell lineage commitment leading to the development of two functionally distinct T cell subsets expressing either TCR-αβ or TCR-γδ heterodimers is unknown. A key question has been whether and how the expressed TCR determines the lineage fate of precursor cells. During the early stages of adult T cell development, the V(D)J recombinase assembles TCR-γ, -δ, and -β genes nearly contemporaneously in the precursor cells that give rise to both γδ and αβ cells. Normal development of the αβ lineage requires the expression of the surrogate α chain (pTα) which associates with a TCR β chain to form the pre-TCR 1. Only thymocytes that express the pre-TCR efficiently traverse a developmental checkpoint at the CD25+CD44− stage of thymocyte development (TCR β chain selection; reference 2). Subsequent differentiation of pre-TCR+ cells to immature CD4+CD8+ thymocytes involves extensive proliferation comprising 8–10 cell divisions 3. Rearrangement of TCR-α genes initiates only after the cells have successfully passed through the TCR β chain selection checkpoint. In contrast, no surrogate chains for γδ lineage development have been identified, and evidence suggests that both chains are required for differentiation along this lineage. Furthermore, γδ T cell development apparently involves much less proliferation than development of αβ lineage cells 4.
Models of T cell lineage commitment vary in the extent to which TCR signals skew the fate decision itself. At one end of the spectrum, the instructive view of γδ/αβ lineage commitment suggests that the γδ TCR and pre-TCR provide distinct signals that stimulate uncommitted precursor cells to differentiate into γδ or αβ lineage cells, respectively 5 6. At the other end of the spectrum is the view that lineage commitment is initially TCR independent, with committed cells able to subsequently rearrange all three relevant TCR genes (γ, δ, and β) 4 6 7 8 9. In this model, TCR signals that are appropriate to the predetermined lineage ensure cell survival and/or expansion, rather than the initial fate choice. Indirect evidence argues against a strict instructive role of the TCR in lineage commitment. For example, in both normal and TCR-β−/− mice, precursor cells expressing γδ TCR can differentiate into CD4+CD8+ αβ lineage cells, albeit inefficiently 4 8. Furthermore, TCR-β expression can lead to the formation of cells with the phenotype of γδ cells 10 11 12. However, the issue remains highly controversial, especially because there has been no direct evidence that T precursor cells, at a stage before TCR gene expression, are divisible into subsets that differ in their potential to differentiate into γδ versus αβ lineage cells.
T cell precursors are contained within a CD4−CD8− thymic subset that does not express the TCR–CD3 complex. This triple negative (TN) population can be further subdivided into four subsets based on cell surface expression of c-kit, CD25, and CD44 molecules, with the following order of maturity: c-kit+CD25−CD44hi (thymic lymphoid precursor) to c-kit+CD25+CD44hi (pro-T) to c-kitloCD25+44− (pre-T) to c-kit−CD25−CD44− 13. TCR-γ, -δ, and -β gene rearrangements occur as cells differentiate from pro-T to pre-T cells 14 15 16. Previous reports 15 16 and our unpublished data indicate that complete (VDJ or VJ) rearrangements of TCR-γ, -δ, and -β alleles are very rare in adult pro-T cells. Quantitative PCR assays demonstrate that <0.5% of pro-T cells contain VJγ or VDJδ rearrangements, while VDJβ rearrangements could not be detected 15. In pre-T cells, 10–75% of TCR alleles are completely rearranged (VDJ or VJ) depending on the TCR locus 16. Hence, the vast majority of CD44hiCD25+ pro-T cells are T lineage precursors lacking functional TCR gene rearrangements. In addition to having T cell developmental potential, pro-T cells can give rise to dendritic cells (DCs), whereas pre-T cells are strictly committed to the T cell lineage 17. In its simplest form, the instructive model of the γδ/αβ lineage commitment predicts that all pro-T cells would be uniform in their developmental potential since they have yet to express functional TCR complexes. In contrast, a TCR-independent mode of lineage commitment can accommodate distinct αβ versus γδ lineage developmental potential in subsets of pro-T cells before TCR expression. As a first step in testing these predictions we show here that in normal young mice, pro-T cell subsets with distinct developmental potential can be defined based on differential expression of the IL-7Rα.
Materials and Methods
Mice.
C57BL/6 (B6), B6-Ly5.1 congenic, B6-G8TCRγ transgenic 18, and B6-IL-7Rα−/− mice (The Jackson Laboratory) were bred and maintained in specific pathogen-free facilities at the University of California at Berkeley.
Antibodies and Flow Cytometry.
Anti–δ TCR (GL-3), anti–β TCR (H57), anti-CD3ε (500A2), anti–IL-7Rα (A7R34), anti-CD45.2 (104), and anti-CD45.1 (A20) mAbs were purified and conjugated with FITC or biotin using standard protocols. Anti-CD44–FITC, anti-CD25–PE, anti–c-kit (CD117)-allophycocyanin (APC), anti-CD8–Tricolor mAbs, and streptavidin-Tricolor were purchased from Caltag. Anti-CD44–Cy5, anti-CD11c–biotin, anti-CD25–FITC, anti-CD45.2 (Ly5.2)–FITC, anti–c-kit–FITC, anti–IL-7Rα (CD127, B12-1)–biotin, anti–mouse I-Ab–PE mAbs, and strepavidin-PE were supplied by BD PharMingen. Anti-CD4–613, anti-CD8–613, anti-CD25–613, and strepavidin-613 were acquired from GIBCO BRL. Rabbit polyclonal antibody against IL-7Rα (D20) was supplied by Santa Cruz Biotechnology, Inc. A7R34 rat anti–mouse IL-7Rα mAb was a gift from Dr. S.-I. Nishikawa and was obtained from Dr. M. Kondo (Stanford University, Palo Alto, CA). Anti–rabbit IgG–biotin and anti–bromodeoxyuridine (BrdU)–FITC were purchased from Vector Laboratories and Becton Dickinson, respectively. Incorporation of BrdU into dividing thymocytes in vivo was carried out by injecting mice intraperitoneally twice at a 2-h interval with 0.9 mg of BrdU 3 4. Thymocytes from injected mice were analyzed 1 h later by flow cytometry. For the analysis of IL-7Rα expression on thymic precursor populations, CD4−CD8− thymocytes were isolated by rabbit and guinea pig complement–mediated lysis of CD4+ and/or CD8+ thymocytes using anti-CD4 (RL172) and CD8 (AD4-15) ascites fluids. After one or two rounds of complement lysis >99% pure double negative (DN) cells were obtained. Cells were incubated with 2.4G2 to block Fc receptors, then stained with mAbs specific for CD25-613, CD44-Cy5, IL-7Rα–biotin/strepavidin-PE, and CD3ε-FITC (or c-kit–FITC), and analyzed by four-color flow cytometry on an XL-MCL flow cytometer (Beckman Coulter). Since virtually all CD25+ DN cells were CD3−, the combination of anti-CD25–FITC/CD44-Cy5/IL-7Rα (B12-1)–biotin-PE and CD8-613 or c-kit–APC mAbs was used for the analysis of pro-T and pre-T subsets. IL-7R+ and IL-7Rneg-lo pro-T cells were sorted with an ELITE cell sorter (Beckman Coulter) using >99% pure DN thymocytes stained with the mAbs as above and then gating on CD44+CD25+ (3% of total DN thymocytes) population and sorting for the cells expressing high levels of IL-7R (25–30% of total pro-T cells) and non- or low expressing cells (20–25% of total pro-T cells). Of the anti–IL-7Rα mAbs, B12-1 gave the best separation and was used for all sorting experiments. Cell yield for each pro-T cell subset was 3–5 × 103 cells/mouse.
Cell Culture, Fetal Thymic Organ Culture, and Intrathymic Injection.
Sorted pro-T cells were cultured for 4 d at 105 cells/ml in medium (RPMI 1640 supplemented with 10% FCS, 50 μM 2-ME, 2 mM l-glutamine, 20 mM Hepes, and antibiotics) containing 10 ng/ml of rIL-7 (Genzyme) or a 1:100 dilution of cell culture supernatants from J558 plasmacytoma cells transfected either with IL-7 or stem cell factor (SCF) cDNAs 19. For fetal thymic organ culture (FTOC), 1.5 × 104 viable, sorted cells were used in a 2-d hanging drop culture to reconstitute fetal day 14 thymic lobe that had been previously depleted of resident thymocytes by treatment with 1.35 mM 2′-deoxyguanosine for 5 d. Repopulated thymi were transferred to a Transwell plate and cultured for 9–24 d. For some experiments, thymi from B6-Ly5.1 mice were used to check for host thymocyte survival in FTOC. Invariably, recipient thymocytes were not detected during FTOC in these experiments. At the early phase of culture (days 9–11) three to five thymi were pooled to obtain sufficient cells for flow cytometric analysis using appropriate mAbs. At a later phase of culture (day 21–22) individual thymi were analyzed. For intrathymic injection, sorted cells from B6 mice were washed and resuspended in PBS/0.1% BSA and injected into thymi of 4–6-wk-old sublethally irradiated (750 rads) B6-Ly5.1 congenic mice. For pro-T cells and pro-T subsets ~2 × 104 sorted viable cells were injected. For pre-T cells ~2 × 105 cells were injected. Reconstituted thymi were analyzed by flow cytometry 8–11 d after injection. DCs were isolated as described 20. In brief, thymi were digested for 1 h at 37°C in serum-free medium containing 1.6 mg/ml collagenase and 0.1% DNase. DCs were incubated with biotinylated anti-CD11c mAb and subsequently with streptavidin-coupled magnetic beads. They were then positively selected by magnetic cell sorting (MACS; Miltenyi Biotec) and analyzed by flow cytometry after staining with Ly5.2 (donor)-FITC and streptavidin-Tricolor that binds to the remaining free biotin of CD11c mAb on DCs.
Reverse Transcription PCR.
RNA samples were reverse transcribed using oligo dT primer and avian reverse transcriptase as described 4. PCRs of serially diluted (three- or fourfold) cDNA and genomic DNA samples were performed using published primers 4 21 in the presence of 1.0 μCi of [α-32P]dCTP. All PCRs entailed 28 cycles except those for TCR-γ or -δ gene rearrangements/transcripts, which entailed 35–38 cycles. The starting input concentration for positive control tubulin PCRs was three- or fourfold lower than the starting concentrations for the gene of interest. Quantitation was performed using a PhosphorImager (Molecular Dynamics).
Results
Nonuniform Expression of IL-7R by Pro-T Cells in Young Mice.
IL-7R–mediated signals are necessary for the survival of the earliest T progenitors 22 23 24, and play an independent, critical role in γδ cell development. In IL-7Rα−/− mice the development of γδ cells is completely abolished 25 26, even when a Bcl2 transgene is expressed 23 24, whereas the development of αβ T cells is blocked incompletely and can be partially rescued by expression of the Bcl2 transgene. The complete block in γδ cell development in IL-7Rα−/− mice was shown to result specifically from a deficiency in TCR γ chain synthesis 18, consistent with the evidence that IL-7Rα signaling stimulates rearrangement 18 27 and expression 18 28 of TCR-γ genes. TCR-δ and TCR-β gene rearrangement and expression are largely unaffected by IL-7Rα deficiency 27.
We examined the expression of IL-7Rα chain on T precursor populations of 3–4-wk-old B6 mice. Similar results were obtained with three different IL-7Rα–specific mAbs (B12-1, A7R34 29, and D20 21, Fig. 1 using B12-1, and data not shown). IL-7Rα was expressed homogeneously at a relatively high level on the earliest thymic T cell progenitors (c-kit+CD25−CD44+ TN). In contrast, pro-T cells (CD25+CD44+c-kit+ TN) exhibited a much broader expression pattern of IL-7Rα chain (Fig. 1A and Fig. B). Using pro-T cells from IL-7Rα−/− mice 22 as a negative staining control, only ~40% of the pro-T cells stained above background, with 15–30% accumulating in the lowest fluorescence intensity channels (Fig. 1 A). Many of the latter cells are not visible in the figure, as they accumulated in the channel corresponding to the left axis of the histogram. At the subsequent pre-T cell stage (c-kitlo CD25+CD44− TN), IL-7Rα was expressed at lower levels than the bright subset of pro-T cells (Fig. 1 A). Four-color cell sorting of TN thymocytes was employed to purify the 25% of pro-T cells expressing the highest levels of IL-7Rα (IL-7Rα+ cells) and the 25% of pro-T cells expressing the lowest levels of IL-7Rα (IL-7Rneg-lo pro-T cells; Fig. 1 B). Both sorted subsets were Thy-1medCD3−CD4−CD8−IL-2Rβ−CD25+CD44+c-kithi (Fig. 1 C, and data not shown). The purity of sorted pro-T subsets ranged from 86 to 95% in 23 independent sorting experiments. The majority (>80%) of sorted IL-7Rlo pro-T thymocytes again accumulated on the lowest fluorescent intensity channel (Fig. 1 C) as a result of the compensation setting used for four-color flow cytometric analysis.
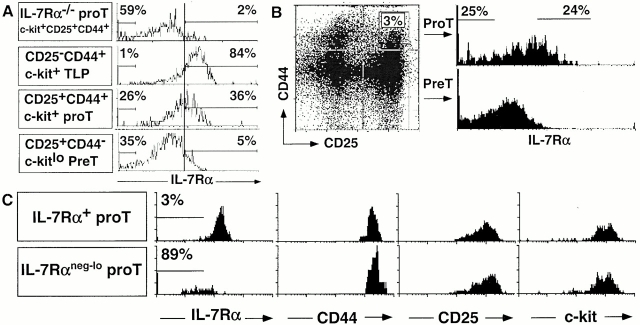
Subdivision of pro-T cells based on IL-7Rα expression. (A) CD4−CD8− thymocytes from B6 and B6 IL-7Rα−/− mice were stained with mAbs for CD25, CD44, IL-7Rα (B12-1), and c-kit. IL-7Rα chain levels on indicated precursor subsets are shown. The level of positive staining for IL-7Rα chain was determined by comparing to the fluorescence signal from IL-7Rα−/− thymocytes (top). An irrelevant, isotype-matched (to B12-1) control mAb did not stain T precursor subsets and CD25+ cells were not stained by mAb specific for CD3ε chain (data not shown). (B) Sorting gates for IL-7R+ and IL-7Rneg-lo CD44+CD25+ TN pro-T cells are shown. IL-7Rα staining of CD25+CD44− TN pre-T cells is presented for comparison. (C) Postsort analysis of similar numbers of purified populations with the indicated mAbs. Sorted cells were additionally stained with anti–c-kit–APC and analyzed on an ELITE flow cytometer. Many of the IL7Rαneg-lo cells in A and C accumulate in the lowest fluorescence intensity channel, characteristic of Beckman Coulter flow cytometers, especially when using four-color compensation settings.
Sorted IL-7Rα+ pro-T cells contained approximately fivefold more IL-7Rα mRNA on average than IL-7Rαneg-lo pro-T cells, based on semiquantitative reverse transcription (RT)-PCR analysis with tubulin mRNA as a control (Fig. 2 A). Expression of the common γ chain (γc) of the IL-2, -4, -7, -9, and -15R complex, on the other hand, was indistinguishable between the two subsets. When cultured for 4 d in the presence of IL-7, sorted IL-7R+ pro-T cells survived better than IL-7Rneg-lo pro-T cells (Fig. 2 B), yielding on average threefold more cells. The difference is due at least in part to a differential responsiveness to IL-7, since the survival of both populations was higher and more similar when a mixture of SCF and IL-7 was used. Analysis of BrdU incorporation during a 3-h pulse demonstrated that the IL-7Rα+ pro-T cell subset proliferates very rapidly in vivo (Fig. 2 C). More of these cells incorporated BrdU than any other thymocyte subset tested under similar conditions 4. On average, only half as many IL-7Rαneg-lo pro-T cells incorporated BrdU in a parallel analysis (Fig. 2 C).
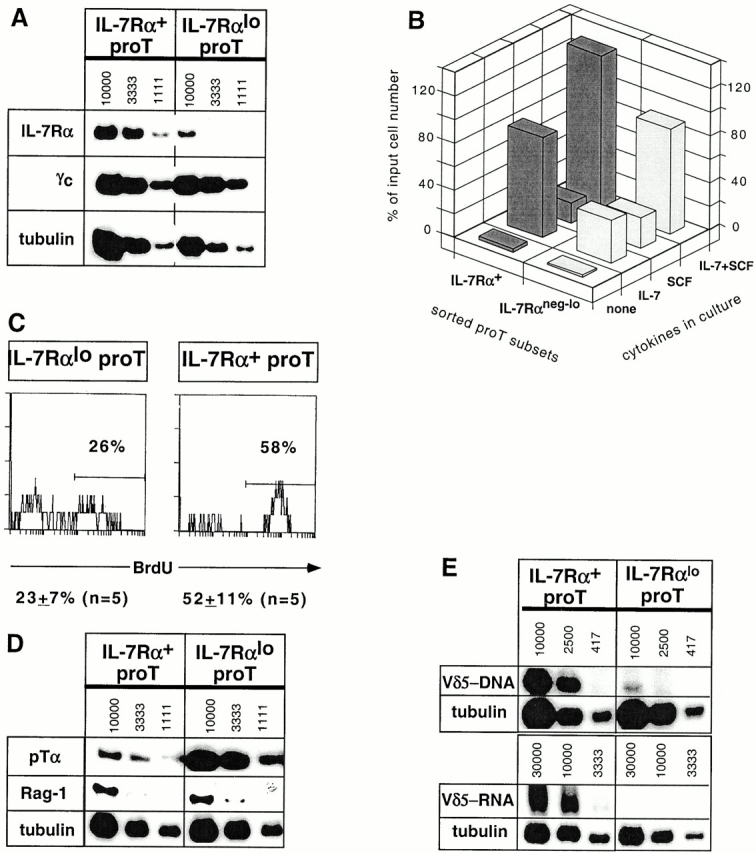
IL-7Rα+ and IL-7Rαneg-lo pro-T subsets exhibit distinct properties. (A) A representative radioactive RT-PCR analysis for IL-7Rα, γc, and tubulin transcripts in the sorted pro-T subsets of similar purity. Numbers indicate approximate cell equivalents used for IL-7Rα and γc-specific PCR. For tubulin PCR, the starting concentration was 3,333 cell equivalents, which was subject to two serial threefold dilutions. The IL-7Rα and γc autoradiographs were exposed for 3 d or overnight, respectively. No PCR products were detected when the RT step was omitted. (B) Proportion of input sorted cells surviving culturing the pro-T subsets for 4 d in the presence of IL-7 and/or SCF. (C) BrdU incorporation after a 3-h pulse with BrdU in vivo. Treated DN thymocytes were stained with mAbs for CD25-613, CD44-Cy5, IL-7Rα-biotin/strepavidin-PE, and BrdU-FITC. The levels of BrdU staining on gated CD25+CD44+IL-7Rα+ and CD25+CD44+IL-7Rαneg-lo TN thymocytes are presented. (D) RT-PCR analysis for pTα and Rag-1 transcripts. Results from one of four independent sorting experiments are shown for pTα expression analysis; two experiments showed a marginal difference (two- to threefold), whereas two others showed a larger (more than ninefold) difference. (E) Levels of Vδ5-Jδ1 rearrangements in genomic DNA (top) and Vδ5-Jδ1 transcripts in total RNA (bottom) from sorted pro-T cell subsets, determined by semiquantitative PCR or RT-PCR, respectively. Genomic DNA PCR was for 35 cycles; RT-PCR entailed 38 cycles for Vδ5-Jδ1 and 28 cycles for tubulin. No PCR products were detected when the RT step was omitted. The autoradiographs shown for Vδ5-Jδ1 and tubulin transcripts were exposed for 5 d and 4 h, respectively.
Semiquantitative RT-PCR analysis demonstrated that IL-7Rα+ and IL-7Rαneg-lo pro-T cells contained similar levels of recombination activating gene (Rag)-1/2 transcripts (Fig. 2 D, and data not shown). Interestingly, expression of the pTα chain, essential for development of most αβ lineage T cells 1, was consistently elevated in IL-7Rαneg-lo pro-T cells than in IL-7Rα+ pro-T cells by an average of fourfold (range: 2–10-fold; Fig. 2 D, and data not shown). Conversely, transcripts corresponding to completely rearranged TCR-δ genes (Fig. 2 E, similar data for Vδ4 not shown) were 6- to >10-fold more abundant in sorted IL-7Rα+ pro-T cells than in IL-7Rαneg-lo pro-T cells. In line with the latter finding, the rare Vδ5-DδJδ rearrangements (<0.2% of alleles 15) were confined almost exclusively to the IL-7Rα+ pro-T cell subset (Fig. 2 E). In contrast, the rare complete Vγ2-Jγ1 rearrangements in the population were equally represented in the two subsets (data not shown). Thus, the IL-7Rαneg-lo subset exhibited properties suggestive of αβ lineage cells whereas rare cells in the IL-7Rα+ subset exhibited at least one characteristic of γδ lineage cells, TCR-δ rearrangement and expression.
IL-7Rα+ and IL-7Rαneg-lo Pro-T Cells Exhibit Differences in Developmental Potential.
The developmental potential of sorted IL-7Rα+ and IL-7Rαneg-lo pro-T cells (from B6-Ly5.2 mice) was examined after intrathymic injection into adult B6-Ly5.1 mice. 7–11 d after engraftment, the reconstituted thymi were analyzed for the content of differentiated γδ+ cells and CD4+CD8+ (αβ lineage) cells that had arisen from donor cells (Fig. 3 A, and Table ). Strikingly, the two pro-T cell populations differed substantially in their lineage potential. In the experiment shown in Fig. 3 A, IL-7Rα+ pro-T cells gave rise to 12-fold more γδ thymocytes than did IL-7Rneg-lo pro-T cells; the average difference was fivefold (Table ). Unseparated pro-T cells exhibited an intermediate capacity to generate γδ cells (Table ). Conversely, IL-7Rneg-lo pro-T cells gave rise to more αβ lineage cells than did IL-7Rα+ cells, by an average of more than fivefold numerically, although the difference was less dramatic when expressed as a percentage of donor-derived cells (Table ). Overall, the average ratio of the percentage of CD4+CD8+ cells and the percentage of γδ+ cells in individual reconstituted mice was >13-fold lower with IL-7Rα+ pro-T cells than with IL-7Rneg-lo pro-T cells (Fig. 3 B). When the host cell types were analyzed in the same animals, the difference in the ratio was limited to twofold (Table ). It is important to note that the sorted donor pro-T cell subpopulations differentiated in the presence of a large excess (20–100-fold; Table ) of developing host thymocytes. Thus, thymic “niches” and other undefined factors that might influence development of the cells should be equivalently available to both populations of injected cells. Therefore, the different outcomes observed indicate that the IL-7Rα+ and IL-7Rneg-lo pro-T cell subpopulations differ intrinsically in their developmental potential.
Table 1
Summary of Intrathymic Injection of T Cell Precursor Subsets
| Donor cells | Host cells | |||
|---|---|---|---|---|
| T precursor cells | γδ | γδ cells | γδ | γδ cells |
| % | ×103 | % | ×103 | |
| IL-7Rα1 pro-T | 5.0 ± 0.7 (11) | 21.8 ± 8.2 (7) | 0.5 ± 0.1 (11) | 124 ± 32 (7) |
| IL-7Rαlo pro-T | 0.9 ± 0.3 (8) | 6.3 ± 1.9 (6) | 0.2 ± 0.0 (8) | 53 ± 12 (6) |
| Pro-T | 1.5 ± 0.4 (5) | 5.0 ± 1.5 (5) | 0.2 ± 0.1 (5) | 59 ± 6 (5) |
| Pre-T | 0.4 ± 0.1 (9) | 2.4 ± 1.4 (8) | 0.6 ± 0.1 (9) | 123 ± 42 (8) |
| PBS/none | 0 (3) | 0 (3) | 0.4 ± 0.0 (3) | 131 ± 22 (3) |
| Donor cells | Host cells | |||
| T precursor cells | CD4+CD8+ | CD4+CD8+ cells | CD4+CD8+ | CD4+CD8+ cells |
| % | ×105 | % | ×105 | |
| IL-7Rα1 pro-T | 54.3 ± 8.2 (11) | 3.0 ± 1.3 (7) | 84.0 ± 4.4 (11) | 267.2 ± 57.4 (7) |
| IL-7Rαlo pro-T | 83.8 ± 6.3 (8) | 14.6 ± 6.7 (6) | 90.8 ± 2.5 (8) | 242.9 ± 41.9 (6) |
| Pro-T | 74.3 ± 8.3 (5) | 3.6 ± 1.0 (5) | 91.0 ± 2.2 (5) | 293.2 ± 50.9 (5) |
| Pre-T | 97.6 ± 0.7 (9) | 12.2 ± 6.0 (8) | 78.9 ± 5.6 (9) | 143.6 ± 26.2 (8) |
| PBS/none | 0 (3) | 0 (3) | 94.7 ± 2.6 (3) | 298.2 ± 30.5 (3) |
Numbers in brackets represent the sample size; cell numbers are averages ± SEM. Significance is versus the corresponding IL-7Rα1 pro-T injected group. Statistics are based on Student's t test.
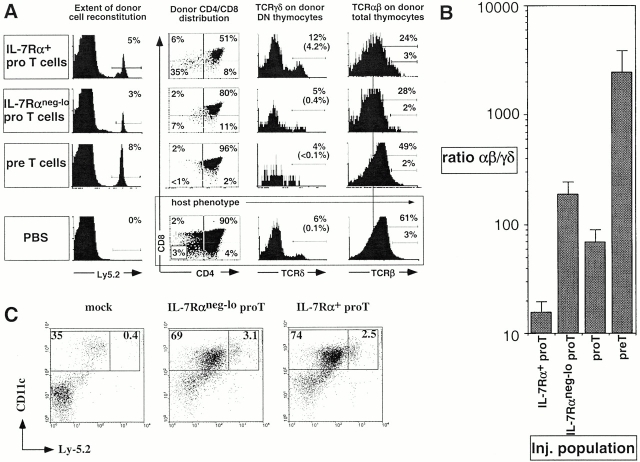
Intrathymically injected pro-T cell subsets displayed distinct γδ versus αβ lineage developmental potential. (A) Representative profiles of thymocytes generated from donor (Ly5.2+) precursor cells. CD4/CD8 profiles of donor-type thymocytes, γδ TCR expression on gated donor-type CD4−CD8− thymocytes, and αβ TCR expression on donor thymocytes are illustrated. Host thymocyte profiles are presented for a mouse injected with saline alone. Percentages in brackets represent the percentage of γδ TCR+ cells of all donor thymocytes. In the profiles compared, IL-7Rα1 and IL-7Rαneg-lo pro-T cells generated similar numbers of donor-derived thymocytes. (B) The ratio of αβ to γδ thymocytes generated from the respective sorted populations 8–11 d after injection. The bars represent averages of αβ/γδ thymocyte ratio of individual mice (see Table ). Total donor thymocyte numbers were higher in the thymi injected with IL-7Rneg-lo pro-T cells, but the broad range (0.2–10.6%) of donor cell reconstitution level in each experimental group makes the interpretation of this difference ambiguous. (C) Representative profiles of five independent experiments show similar proportions of DCs generated from the pro-T subsets (average proportions of donor DCs in CD11c+ population ± SEM: IL-7Rαneg-lo, 10.4 ± 2.6%; IL-7Rα+, 12.2 ± 2.4%). Thymic DCs were purified from mice 9 d after intrathymic injection of pro-T cells or no cells (mock), as indicated. The cells were analyzed for CD11c expression, which identifies all DCs, and for Ly-5.2 expression, which distinguishes DCs of donor (Ly-5.2+) versus host (Ly-5.2−) origin. Purified DCs (all MHC class II+) from mice with 4% and 1.5–1.9% donor cell reconstitution from injected IL-7Rαneg-lo and IL-7Rα+ pro-T cells, respectively, are shown. For comparison, cells from a mouse that underwent the same surgical procedure but was not injected are presented (mock).
Intrathymically injected pre-T cells efficiently differentiated into αβ lineage cells, but yielded extremely few γδ cells (Table ; Fig. 3A and Fig. B). In some experiments pre-T cells yielded no detectable γδ T cells. Hence the αβ/γδ ratio for pre-T cells was 10-fold higher than that of IL-7Rneg-lo pro-T cells, and 100-fold higher than IL-7Rα+ pro-T cells. The results suggest that most pre-T cells are restricted to the αβ lineage. It should also be noted that ~10 times as many pre-T cells as IL-7Rneg-lopro-T cells had to be injected to generate similar numbers of αβ lineage cells (Table ). The much lower cell generative capacity of pre-T cells compared with pro-T cells excludes the possibility that contaminating pre-T cells can account for the αβ lineage preference of the IL-7Rneg-lo pro-T cell subset.
In contrast to the apparent γδ/αβ lineage bias in pro-T subsets, injected IL-7Rα+ and IL-7Rαneg-lo pro-T cell subsets, like unseparated pro-T cells 17, generated similar proportions of lymphoid DCs (Ly5.2 donor type, MHC class II+ and CD11c+; Fig. 3 C). Thus, the two pro-T subsets do share some developmental properties associated with an early T progenitor population, but specifically differ in their ability to generate γδ versus αβ lineage cells.
The lineage bias observed in the pro-T subsets developing in the adult thymic environment in vivo was recapitulated in FTOC. Host thymocyte-depleted E14 thymi were repopulated with sorted pro-T cells. At a relatively early stage of the cultures (days 9–11), IL-7Rα+ pro-T cells yielded approximately three times the percentage of γδ thymocytes as IL-7Rαneg-lo pro-T cells (Fig. 4 A and Table ). Conversely, IL-7Rα+ pro-T cells generated fewer αβ lineage cells than IL-7Rαneg-lo cells, although the difference was not striking. The total numbers of thymocytes generated from the two pro-T subsets in FTOC were not significantly different (Table ). Thus, as was observed in the intrathymic injection studies, the average ratio of αβ/γδ lineage cells generated by IL-7Rαneg-lo pro-T cells was higher than that of IL-7Rα+ pro-T cells, in this case by a factor of five on average (Table ). At a later stage of FTOC (21–22 d) both input populations yielded somewhat lower percentages of γδ cells and higher percentages of αβ lineage cells, probably because of greater expansion of αβ lineage cells. Nevertheless, the IL-7Rαneg-lo pro-T cells still yielded fewer γδ cells and more αβ lineage cells than IL-7Rα+ pro-T cells, resulting in an average threefold difference in the αβ/γδ ratio (Table ). Thus, the lineage bias of the two pro-T cell subsets is apparent at both early and late stages of thymic reconstitution.
Table 2
Summary of FTOC of T Cell Precursor Subsets
| Day of culture | T precursor cells | n | Total cells | γδ | γδ cells | CD4+CD8+ | CD4+CD8+ cells |
|---|---|---|---|---|---|---|---|
| ×103 | % | ×103 | % | ×103 | |||
| Day 9–11 | Il-7Rα1 pro-T | 14 | 26.1 ± 3.7 | 21.1 ± 3.0 | 5.8 ± 1.2 | 26.8 ± 7.1 | 6.8 ± 1.9 |
| IL-7Rαlo pro-T | 11 | 37.2 ± 2.6 | 7.8 ± 0.4 | 2.9 ± 0.3 | 34.1 ± 5.1 | 12.8 ± 2.2 | |
| ProT | 4 | 13.6 | 12.3 | 1.7 | 16.2 | 2.2 | |
| PreT | 4 | 34.7 | 1.1 | 0.4 | 61.4 | 23.1 | |
| TG+IL-7Rα+ pro-T | 7 | 14.6 ± 2.4 | 64.0 ± 5.7 | 9.2 ± 0.7 | 15.2 ± 2.8 | 2.3 ± 0.8 | |
| TG+IL-7Rαlo pro-T | 8 | 20.0 ± 1.2 | 25.4 ±2.1 | 5.1 ± 0.1 | 30.2 ± 2.0 | 6.0 ± 0.7 | |
| Day 21/22 | IL-7Rα1 pro-T | 12 | 40.6 ± 3.5 | 11.6 ± 0.5 | 4.7 ± 0.4 | 44.2 ± 3.4 | 18.1 ± 2.4 |
| IL-7Rαlo pro-T | 12 | 46.8 ± 3.0 | 3.5 ± 0.2 | 1.6 ± 0.1 | 62.1 ± 3.0 | 29.1 ± 2.1 |
TG, G8 TCR-γ transgenic; cell numbers/lobes are averages ± SEM; for pro-T and pre-T cultures, results represent analysis of four pooled thymic lobes. Significance is versus the corresponding IL-7Rα1 pro-T injected group. Statistics are based on the Student's t test.
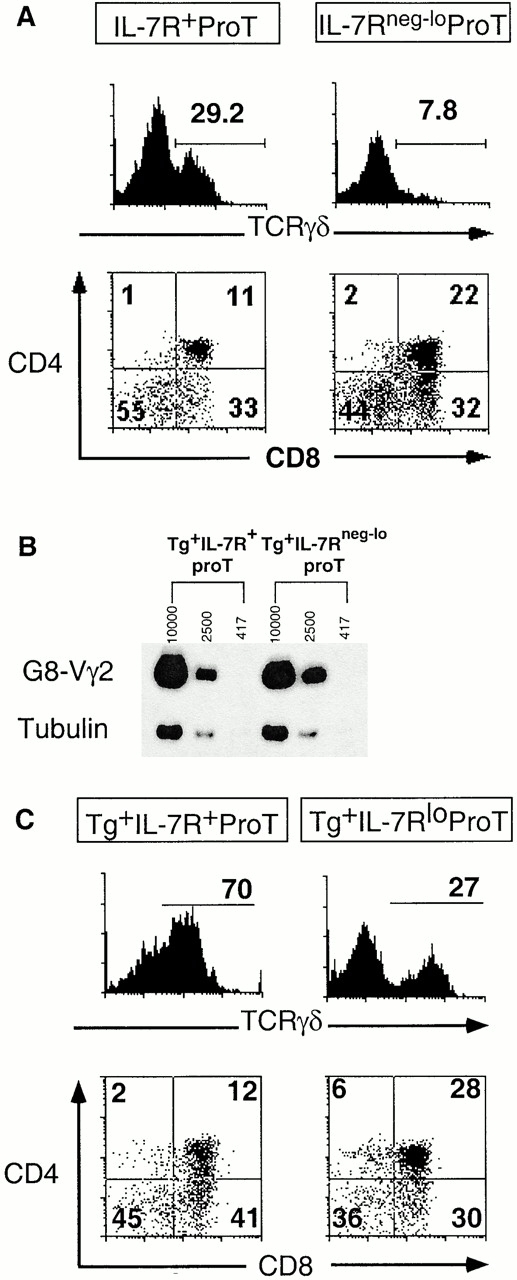
The sorted pro-T subsets display distinct γδ/αβ lineage developmental potential in FTOC. (A) Thymocytes from FTOCs reconstituted with sorted pro-T cell subsets were analyzed for γδ TCR and CD4/CD8 expression after 11 d of culture. Representative profiles are from three thymic lobes that were pooled. Many of the cells are accumulating on the axes of the profiles as a result of four-color compensation settings. (B) Similar transgene expression in sorted pro-T cell subsets from low 1 2 transgene copy line as determined by semiquantitative RT-PCR using transgene-specific primers. Numbers indicate approximate cell equivalents. Two additional experiments yielded comparable results. No PCR products were detected when the RT step was omitted. (C) Thymocytes from FTOCs reconstituted with sorted pro-T cell subsets from high copy G8 transgenic mice were analyzed for γδ TCR and CD4/CD8 expression after 10 d of culture. The levels of IL-7Rα expression on the pro-T progenitor populations in the transgenic mice were not notably different from those in nontransgenic littermates. Three thymic lobes of each type were pooled for analysis.
The Two Subsets of Pro-T Cells Support TCR-γ Expression Equally.
It was possible that the IL-7Rα+ population generates more γδ cells simply because the higher IL-7Rα levels enhance responsiveness of the cells to IL-7, leading to augmented rearrangement and/or transcription of TCR-γ genes. Semiquantitative PCR data indicated, however, that TCR-γ rearrangements were equally prevalent in the two pro-T cell subsets, though very rare in both cases (data not shown). Since most cells in both populations did not contain rearranged TCR-γ alleles, an alternative approach was developed to assess whether the subsets differ in the capacity to express rearranged TCR-γ genes. A previous study of mice with a prerearranged TCR-γ transgene present at low copy number (<2) demonstrated that transgene transcription was reduced by 26-fold when the mice lacked a functional IL-7Rα gene 18. These data suggest that transgene transcription is an excellent assay for IL-7Rα–dependent TCR-γ transcription. Semiquantitative RT-PCR analysis revealed that the TCR-γ transgene was transcribed equally in IL-7Rα+ and IL-7Rαneg-lo pro-T cell subsets (Fig. 4 B). This result suggests either that the low IL-7Rα level on IL-7Rneg-lo pro-T cells is sufficient to stimulate TCR-γ gene rearrangement and transcription, or that IL-7Rα signaling at the earlier c-kit+CD25−CD44+ stage activates the TCR-γ locus in a manner that is sustained in later stages. Most significantly, the data suggest that despite having different levels of IL-7Rα, the two subsets do not differ in the capacity to express TCR-γ genes.
Another approach was employed to address the role of IL-7R–dependent TCR-γ rearrangement/transcription. If the distinct developmental potentials of the two pro-T cell subsets was due simply to differential expression and/or rearrangement of TCR-γ genes, the difference should be abolished by equipping both subsets with a TCR-γ gene that is expressed adequately even in the absence of IL-7Rα signaling. Previous analysis of a high copy (~33 copies) TCR-γ transgenic line crossed into an IL-7Rα−/− background showed that the transgene is expressed at a higher level than endogenous TCR-γ genes are in normal mice 18. The high number of transgene copies compensates for reduced transcription of each copy, resulting in a high aggregate transcription level. Importantly, this level of transcription was shown to be sufficient to restore development of γδ T cells in IL-7Rα−/− mice. As expected from the previous results with the low copy transgene, the high copy transgene was expressed equally in sorted IL-7Rα+ and IL-7Rneg-lo pro-T cells (data not shown). As assayed in FTOCs, both subsets yielded more γδ cells than wild-type subsets, presumably because the in-frame transgene provides functional γ chains to all cells whereas productive γ rearrangements occur in only a fraction of cells in nontransgenic mice. However, the high copy transgene did not equalize the developmental potential of the pro-T cell subsets as assayed in FTOCs (Fig. 4 C, and Table ). The IL-7Rα+ pro-T cells yielded an average of 2.6 times the percentage of γδ cells and half the percentage of αβ lineage cells as did IL-7Rneg-lo pro-T cells. The collective data indicate that the two subsets exhibit distinct developmental potential even when TCR-γ expression is driven in all cells at higher than normal levels, and further, that the two subsets do not differ in the extent of TCR-γ rearrangement or the capacity to support TCR-γ expression. Clearly, other differences between the subsets must account for their distinct developmental capacities.
Discussion
We present data demonstrating that pro-T cells are heterogeneous in phenotype and that the subpopulations of pro-T cells do not generate equivalent progenies. A trivial explanation of the data would be that the sorted IL-7Rαneg-lo subset was selectively contaminated with more mature IL-7Rαneg pre-T cells. Our data indicate that the latter cells exhibit a strong bias to the αβ lineage (Fig. 3 B). This possibility is highly unlikely, however, because pre-T cells in the intrathymic assay exhibited ~10-fold less cell generative capacity compared with IL-7Rαneg-lo pro-T cells (Table ) and would therefore not be capable of generating sufficient numbers of αβ lineage cells to account for the results. For example, after intrathymic injection of low numbers of pre-T cells (~104), equivalent to half the number of pro-T cells that were injected, negligible progeny were detected (data not shown). We therefore consider various substantive explanations for the observed lineage bias of pro-T cell subsets.
Are the Pro-T Cell Subsets Sequentially Related Populations That Exhibit Distinct TCR Rearrangement Potential, but No Other Intrinsic Lineage Bias?
One possibility is that the IL-7Rα+ pro-T cell population precedes the IL-7Rαneg-lo population in a developmental sequence. If cells at the IL-7Rα+ stage rearrange TCR-γ and -δ genes in preference to TCR-β genes, the population would exhibit a relative bias in favor of γδ cell development. Failure to successfully initiate γδ lineage development at this stage would result in subsequent differentiation of IL-7Rαneg-lo pro-T cells. At this stage TCR-γ, -δ, and -β genes may rearrange more or less equivalently. Equivalent rearrangement of the three genes would marginally favor αβ lineage development, which requires only one in-frame rearrangement (of TCR-β) as opposed to the two in-frame rearrangements (of γ and δ) required for γδ lineage development. This scheme differs substantially from models in which the two populations represent divergent paths, though it does imply different lineage potential in the two populations due to an early preference for TCR-γ and -δ rearrangements.
Several lines of evidence are inconsistent with this sequential model. First, if all developing T cells traverse the IL-7Rα+ pro-T cell stage, the latter cells should ultimately yield αβ and γδ lineage progeny in physiological proportions, i.e., a ratio of 100 or more, and should not exhibit a significant bias for γδ lineage development. In fact, the IL-7Rα+ pro-T cell subset yielded an αβ/γδ ratio of ~15 (Fig. 3 B). The discrepancy is unlikely to reflect differences in developmental kinetics in the populations, because the lineage bias was evident at both early and late times after initiation of FTOCs (Table ). In contrast, unseparated pro-T cells yielded αβ and γδ progeny in physiological proportions. A second persuasive argument against the sequential scheme arises from its prediction that many cells in the IL-7Rαneg-lo population should have rearrangements at the TCR-δ and/or -γ loci. This is because the majority (67%) of the TCR-γ and -δ rearrangements that would occur at the “early” IL-7Rα+ stage are predicted to be nonproductive. Many cells with productive rearrangements of one of the genes (γ or δ) would have nonproductive rearrangements of the other, and some cells would have only nonproductive rearrangements at both loci. All of these cells would be unable to differentiate into γδ cells. If they have no intrinsic lineage bias they should then convert to the “subsequent” IL-7Rαneg-lo population, contributing numerous chromosomes with rearranged TCR-γ and -δ alleles. In strong contrast to this expectation, the latter population is essentially devoid of cells with TCR-δ rearrangements (Fig. 2 E), and contains a very low frequency of rearranged TCR-γ alleles similar to that in the IL-7Rα+ population. Yet, αβ lineage cells are known to contain high levels of TCR-γ and -δ rearrangements, most of which are nonproductive 30 31 32. It is likely that these rearrangements occur subsequent to the IL-7Rαneg-lo stage rather than before. These data argue against the proposal that IL-7Rα+ pro-T cells preferentially undergo TCR-γ and -δ rearrangements before differentiating into IL-7Rαneg-lo pro-T cells. Although the results do not rule out all conceivable models in which the two subsets are sequentially related, the simplest interpretation is that the two subsets represent alternative, as opposed to sequential, developmental stages.
Is Differential IL-7R Signaling in Pro-T Cells Responsible for the Observed Lineage Bias?
Previous reports demonstrate that IL-7R signaling plays a unique role in γδ cell development, as it promotes rearrangement and expression of TCR-γ genes but is not necessary for rearrangement or expression of the other TCR genes 18 27 28 33. Thus, an obvious possibility was that the bias of the IL-7Rα+ subset for γδ lineage development resulted from enhanced rearrangement and expression of TCR-γ genes in this subset. Conversely, the αβ lineage bias of the IL-7Rαneg-lo pro-T cell subset might be because of impaired rearrangement and expression of TCR-γ genes. However, several lines of evidence indicate that IL-7Rαneg-lo pro-T cells and pre-T cells have the capacity to support high levels of TCR-γ gene expression, despite the low or absent levels of IL-7Rs on these cells. First, the low copy prerearranged TCR-γ transgene, whose transcription was previously shown to be IL-7Rα dependent, was expressed at a similar level in the pro-T subsets (Fig. 4 B). Second, in pre-T cells, endogenous TCR-γ rearrangements are abundant, and these are expressed at a high level despite negligible IL-7R on these cells (data not shown). Third, Vγ2-Jγ1 rearrangements, although very rare at the pro-T cell stage, are equally represented in the two pro-T cell subpopulations as determined by semiquantitative PCR (data not shown). Finally, it is notable that αβ lineage cells contain very high levels of mostly nonproductive TCR-γ rearrangements 34 35; it is therefore implausible to argue that rearrangement of TCR-γ genes is impaired in the progenitors of these cells. Collectively these data suggest either that IL-7Rα signaling at an earlier stage of development allows TCR-γ gene activation at a later stage, or that low levels of IL-7R are sufficient to activate the locus at the pro- and pre-T cell stages.
Although these arguments suggest that the two pro-T cell subsets are equally capable of activating the TCR-γ locus, we sought a more direct test of whether the lineage bias was attributable to impaired TCR-γ gene expression. The results indicated that the lineage bias was still evident even in the presence of high levels of functional TCR γ chain in both pro-T subsets, directed by a high copy transgene (Fig. 4 C). These findings represent a strong argument against the possibility that the lineage bias of the pro-T cell subsets results from differential TCR-γ gene activation as a consequence of distinct IL-7Rα levels. Nevertheless, IL-7Rα levels correlate with lineage-biased subsets in young adult mice. It should be noted that pro-T cells in newborn mice uniformly express high levels of IL-7Rα (data not shown). Thus, the correlation between IL-7R levels on pro-T cells and lineage potential may hold true only in a homeostatic steady-state thymic environment.
Are Pro-T Cells Committed to Developing into γδ or αβ Cells before TCR Gene Rearrangement?
The selective differences in αβ/γδ composition, but not in DCs, in the progenies of the pro-T subsets appear to be most consistent with the possibility that pro-T cells already contain γδ and αβ lineage–biased precursor cells. This lineage bias is TCR independent since a negligible fraction of pro-T cells express functional TCR. It should be noted that even the relatively γδ-biased IL-7Rα+ population generated more αβ lineage cells than γδ cells, probably because αβ lineage cells undergo 8–10 rounds of cell division 3, whereas γδ lineage cells undergo much less expansion 4. Thus, the IL-7Rα+pro-T cell population probably contains fewer αβ lineage precursors than γδ lineage precursors, with subsequent expansion of the αβ lineage cells eventually leading to an excess of these cells.
A caveat of our results is that the two sorted populations appear to be incompletely restricted in their lineage potential. Subsets defined by differential IL-7R expression generate cells of both αβ and γδ lineages, albeit at different relative ratios. The precursor cells may be only biased rather than completely restricted in their lineage potential, but other possibilities must be considered. For example, the IL-7Rα marker may not precisely define restricted precursor cells, or the cell sorting may have been imperfect. It is also possible that an initial bias in lineage potential is subject to modification by signals occurring later in development, including signals through the TCR. Nevertheless, our data do argue strongly against a simple instructive model in which the lineage fate decision of T cell precursors is determined primarily by which TCR, γδ, or pre-TCR is expressed. Obviously, a further refinement of our understanding of T cell lineage commitment at the pro-T cell level awaits in vivo clonal analyses of developmental potential of single pro-T cells sorted from adult mice.
We propose that a thymic lymphoid progenitor generates two distinct lineage-biased populations at or before the pro-T cell stage. Most γδ lineage–committed precursor cells that express a functional TCR-γδ will develop into γδ lineage cells whether or not they have previously or concomitantly expressed TCR β chain, consistent with published data 10 11. Conversely, αβ lineage–committed precursor cells that express the pre-TCR will proliferate and give rise to a predominant population of double positive thymocytes 3. It has been demonstrated that αβ lineage–biased precursors that express functional γδ TCRs instead of the pre-TCR can differentiate into double positive thymocytes 4 8. However, TCR-γ and -δ gene expression is then extinguished in these cells 4 36 37, presumably leading to their eventual death. Hence, although lineage commitment may be initiated well before functional TCR expression, subsequent expression of the corresponding type of TCR is normally necessary for the ultimate maturation of the cells.
Acknowledgments
We thank Drs. C. Chambers, E. Robey, L. Berg, and R. Vance for helpful discussions and review of the manuscript; P. Schow, H. Nolla, R. Vance, and C. McMahon for technical assistance with flow cytometric cell sorting; and A. Lazar for providing B6 mice.
This work was supported by grants to D.H. Raulet from the National Institutes of Health. J. Kang was a research fellow of the National Cancer Institute of Canada supported by the Canadian Cancer Society. A. Volkmann was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation.
Footnotes
Abbreviations used in this paper: APC, allophycocyanin; BrdU, bromodeoxyuridine; B6, C57BL/6; DC, dendritic cell; DN, double negative; FTOC, fetal thymic organ culture; RT, reverse transcription; SCF, stem cell factor; TN, triple negative.
References
- Fehling H., Krotkova A., Saint-Ruf C., von Boehmer H. Crucial role of the pre-T cell receptor α gene in the development of αβ but not γδ T cells. Nature. 1995;375:795–798. [Abstract] [Google Scholar]
- Dudley E.C., Petrie H.T., Shah L.M., Owen M.J., Hayday A.C. T cell receptor β chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1994;1:83–93. [Abstract] [Google Scholar]
- Penit C., Lucas B., Vasseur F. Cell expansion and growth arrest phases during the transition from precursor (CD4−8−) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J. Immunol. 1995;154:5103–5113. [Abstract] [Google Scholar]
- Kang J., Coles M., Cado D., Raulet D.H. The developmental fate of T cells is critically influenced by TCR γδ expression. Immunity. 1998;8:427–438. [Abstract] [Google Scholar]
- Allison J.P., Lanier L.L. The T-cell antigen receptor γ generearrangement and cell lineages. Immunol. Today. 1987;8:293–296. [Abstract] [Google Scholar]
- Kang J., Raulet D.H. Events that regulate differentiation of αβ TCR+ and γδ TCR+ cells from a common precursor. Semin. Immunol. 1997;9:171–179. [Abstract] [Google Scholar]
- Winoto A., Baltimore D. Separate lineages of T cells expressing the αβ and γδ receptors. Nature. 1989;338:430–432. [Abstract] [Google Scholar]
- Livak F., Wilson A., MacDonald H.R., Schatz D.G. αβ lineage-committed thymocytes can be rescued by the γδ T cell receptor (TCR) in the absence of TCR β chain. Eur. J. Immunol. 1997;27:2948–2958. [Abstract] [Google Scholar]
- MacDonald H.R., Wilson A. The role of the T-cell receptor (TCR) in αβ/γδ lineage commitmentclues from intracellular TCR staining. Immunol. Rev. 1998;165:87–94. [Abstract] [Google Scholar]
- Bruno L., Fehling H.J., von Boehmer H. The αβ T cell receptor can replace the γδ receptor in the development of γδ lineage cells. Immunity. 1996;5:343–352. [Abstract] [Google Scholar]
- Wilson A., MacDonald H.R. A limited role for β-selection during γδT cell development. J. Immunol. 1998;161:5851–5854. [Abstract] [Google Scholar]
- Terrence K., Pavlovich C.P., Matechak E.O., Fowlkes B.J. Premature expression of T cell receptor (TCR)αβ suppresses TCRγδ gene rearrangement but permits development of γδ lineage t cells. J. Exp. Med. 2000;192:537–548. [Europe PMC free article] [Abstract] [Google Scholar]
- Godfrey D., Zlotnik A. Control points in early T-cell development. Immunol. Today. 1993;14:547–553. [Abstract] [Google Scholar]
- Godfrey D.I., Kennedy J., Mombaerts P., Tonegawa S., Zlotnik A. Onset of TCR-β gene rearrangement and role of TCR-β expression during CD3−CD4−CD8− thymocyte differentiation. J. Immunol. 1994;152:4783–4792. [Abstract] [Google Scholar]
- Capone M., Hockett R.D., Zlotnik A. Kinetics of T cell receptor β, γ, and δ rearrangements during adult thymic developmentT cell receptor rearrangemnts are present in CD44+CD25+ pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 1998;95:12522–12527. [Europe PMC free article] [Abstract] [Google Scholar]
- Livák F., Tourigny M., Schatz D.G., Petrie H.T. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 1999;162:2575–2580. [Abstract] [Google Scholar]
- Wu L., Li C.L., Shortman K. Thymic dendritic cell precursorsrelationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 1996;184:903–911. [Europe PMC free article] [Abstract] [Google Scholar]
- Kang J., Coles M., Raulet D.H. Defective development of γδ T cells in interleukin 7 receptor-deficient mice is due to impaired expression of T cell receptor γ genes. J. Exp. Med. 1999;190:973–982. [Europe PMC free article] [Abstract] [Google Scholar]
- Karasuyama H., Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur. J. Immunol. 1988;18:97–104. [Abstract] [Google Scholar]
- Volkmann A., Zal T., Stockinger B. Antigen-presenting cells in the thymus that can negatively select MHC class II-restricted T cells recognizing a circulating self antigen. J. Immunol. 1997;158:693–706. [Abstract] [Google Scholar]
- Hikida M., Nakayama Y., Yamashita Y., Kumazawa Y., Nishikawa S.I., Ohmori H. Expression of recombination activating genes in germinal center B cellsinvolvement of interleukin 7 (IL-7) and the IL-7 receptor. J. Exp. Med. 1998;188:365–372. [Europe PMC free article] [Abstract] [Google Scholar]
- Peschon J.J., Morrissey P.J., Grabstein K.H., Ramsdell F.J., Maraskovsky E., Gliniak B.C., Park L.S., Ziegler S.F., Williams D.E., Ware C.B. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J. Exp. Med. 1994;180:1955–1960. [Europe PMC free article] [Abstract] [Google Scholar]
- Akashi K., Kondo M., von Freeden-Jeffry U., Murray R., Weissman I.L. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. [Abstract] [Google Scholar]
- Maraskovsky E., O'Reilly L.A., Teepe M., Corcoran L.M., Peschon J.J., Strasser A. Bcl-2 can rescue T lymphocyte development in interlukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89:1011–1019. [Abstract] [Google Scholar]
- He Y.W., Malek T.R. Interleukin-7 receptor α is essential for the development of γδ1 T cells, but not natural killer cells. J. Exp. Med. 1996;184:289–293. [Europe PMC free article] [Abstract] [Google Scholar]
- Maki K., Sunaga S., Komagata Y., Kodaira Y., Mabuchi A., Karasuyama H., Yokomuro K., Miyazaki J.I., Ikuta K. Interleukin 7 receptor-deficient mice lack γδ T cells. Proc. Natl. Acad. Sci. USA. 1996;93:7172–7177. [Europe PMC free article] [Abstract] [Google Scholar]
- Maki K., Sunaga S., Ikuta K. The V-J recombination of T cell receptor γ genes is blocked in interleukin-7 receptor–deficient mice. J. Exp. Med. 1996;184:2423–2427. [Europe PMC free article] [Abstract] [Google Scholar]
- Perumal N.B., Kenniston T.W., Jr., Tweardy D.J., Dyer K.F., Hoffman R., Peschon J., Appasamy P.M. TCR-γ genes are rearranged but not transcribed in IL-7R α-deficient mice. J. Immunol. 1997;158:5744–5750. [Abstract] [Google Scholar]
- Sudo T., Nishikawa S., Ohno N., Akiyama N., Tamakoshi M., Yoshida H. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. USA. 1993;90:9125–9129. [Europe PMC free article] [Abstract] [Google Scholar]
- Dudley E.C., Girardi M., Owen M.J., Hayday A.C. αβ and γδ T cells can share a late common precursor. Curr. Biol. 1995;5:659–669. [Abstract] [Google Scholar]
- Kang J., Baker J., Raulet D. Evidence that productive rearrangements of TCRγ genes influence the fate of developing T cells. Eur. J. Immunol. 1995;25:2706–2709. [Abstract] [Google Scholar]
- Livak F., Petrie H.T., Crispe I.N., Schatz D.G. In-frame TCR δ rearrangements play a critical role in the αβ/γδ lineage decision. Immunity. 1995;2:617–627. [Abstract] [Google Scholar]
- Schlissel M.S., Durum S.D., Muegge K. The interleukin 7 receptor is required for T cell receptor γ locus accessibility to the V(D)J recombinase. J. Exp. Med. 2000;191:1045–1050. [Europe PMC free article] [Abstract] [Google Scholar]
- Kranz D.M., Saito H., Heller M., Takagaki Y., Haas W., Eisen H.N., Tonegawa S. Limited diversity of the rearranged T-cell γ gene. Nature. 1985;313:752–755. [Abstract] [Google Scholar]
- Garman R.D., Doherty P.J., Raulet D.H. Diversity, rearrangement and expression of murine T cell γ genes. Cell. 1986;45:733–742. [Abstract] [Google Scholar]
- Ishida I., Verbeek S., Bonneville M., Itohara S., Berns A., Tonegawa S. T-cell receptor γδ and γ transgenic mice suggest a role of a γ gene silencer in the generation of αβ T cells. Proc. Natl. Acad. Sci. USA. 1990;87:3067–3071. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilson A., de Villartay J.P., MacDonald H.R. T cell receptor δ gene rearrangement and T early α (TEA) expression in immature αβ lineage thymocytesimplications for αβ/γδ lineage commitment. Immunity. 1996;4:37–45. [Abstract] [Google Scholar]
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.193.6.689
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jem/article-pdf/193/6/689/1134364/001716.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1084/jem.193.6.689
Article citations
Harnessing the Power of IL-7 to Boost T Cell Immunity in Experimental and Clinical Immunotherapies.
Immune Netw, 24(1):e9, 15 Feb 2024
Cited by: 2 articles | PMID: 38455462
Review
Skin γδ T Cells and Their Function in Wound Healing.
Front Immunol, 13:875076, 11 Apr 2022
Cited by: 22 articles | PMID: 35479079 | PMCID: PMC9035842
Review Free full text in Europe PMC
γδ T Cells in Skin Inflammation.
Crit Rev Immunol, 42(5):43-56, 01 Jan 2022
Cited by: 2 articles | PMID: 37075018 | PMCID: PMC10439530
Review Free full text in Europe PMC
αβ/γδ T cell lineage outcome is regulated by intrathymic cell localization and environmental signals.
Sci Adv, 7(29):eabg3613, 14 Jul 2021
Cited by: 2 articles | PMID: 34261656 | PMCID: PMC8279519
Gamma Delta T Cells and Their Pathogenic Role in Psoriasis.
Front Immunol, 12:627139, 25 Feb 2021
Cited by: 35 articles | PMID: 33732249 | PMCID: PMC7959710
Review Free full text in Europe PMC
Go to all (77) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Premature expression of T cell receptor (TCR)alphabeta suppresses TCRgammadelta gene rearrangement but permits development of gammadelta lineage T cells.
J Exp Med, 192(4):537-548, 01 Aug 2000
Cited by: 99 articles | PMID: 10952723 | PMCID: PMC2193230
αβ versus γδ fate choice: counting the T-cell lineages at the branch point.
Immunol Rev, 238(1):169-181, 01 Nov 2010
Cited by: 37 articles | PMID: 20969592 | PMCID: PMC3031133
Review Free full text in Europe PMC
Human alpha beta and gamma delta thymocyte development: TCR gene rearrangements, intracellular TCR beta expression, and gamma delta developmental potential--differences between men and mice.
J Immunol, 176(3):1543-1552, 01 Feb 2006
Cited by: 40 articles | PMID: 16424183 | PMCID: PMC1592528
Lineage divergence at the first TCR-dependent checkpoint: preferential γδ and impaired αβ T cell development in nonobese diabetic mice.
J Immunol, 186(2):826-837, 10 Dec 2010
Cited by: 10 articles | PMID: 21148803 | PMCID: PMC4087166





