Abstract
Free full text

Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding
Abstract
The DNA-binding domain (DBD) of progesterone receptor (PR) is bipartite containing a zinc module core that interacts with progesterone response elements (PRE), and a short flexible carboxyl terminal extension (CTE) that interacts with the minor groove flanking the PRE. The chromosomal high-mobility group B proteins (HMGB), defined as DNA architectural proteins capable of bending DNA, also function as auxiliary factors that increase the DNA-binding affinity of PR and other steroid receptors by mechanisms that are not well defined. Here we show that the CTE of PR contains a specific binding site for HMGB that is required for stimulation of PR-PRE binding, whereas the DNA architectural properties of HMGB are dispensable. Specific PRE DNA inhibited HMGB binding to the CTE, indicating that DNA and HMGB–CTE interactions are mutually exclusive. Exogenous CTE peptide increased PR-binding affinity for PRE as did deletion of the CTE. In a PR-binding site selection assay, A/T sequences flanking the PRE were enriched by HMGB, indicating that PR DNA-binding specificity is also altered by HMGB. We conclude that a transient HMGB–CTE interaction alters a repressive conformation of the flexible CTE enabling it to bind to preferred sequences flanking the PRE.
INTRODUCTION
Progesterone receptor (PR) is a member of the nuclear receptor superfamily, which is composed of ligand-dependent transcription factors that regulate a variety of cellular processes including metabolism, development, growth, differentiation and reproduction. Transcriptional regulation occurs through direct receptor binding to specific hormone response elements (HREs) of target genes. For steroid receptors, optimized HREs are hexanucleotide sequences arranged as inverted repeats with a trinucleotide spacer (3N) between the half sites (1–3). The DNA-binding domain (DBD) of nuclear receptors is bipartite, consisting of a core domain and an adjacent ~30–40-amino acid segment termed the C-terminal extension (CTE). The core of the DBD is a highly conserved structure that mediates base-specific contacts in the major groove of the HRE DNA (1,3,4). The CTE is a nonconserved flexible region that adopts different structures depending on the specific receptor and it interacts with the DNA minor groove outside the HRE (5–16).
In the structures of class II nuclear receptor DBDs, such as thyroid receptor, (TR), (6,8,9,14) retinoic acid receptor (RAR) and RXR, the CTE formed a single α-helix (termed A box) that projects across the DNA minor groove and makes extensive nonspecific contacts with the DNA backbone. The CTEs of orphan receptors, such as RevErb, NGF1-B and ERR-2, bind to the minor groove in an extended loop conformation (5,7,13,16). The ERR-2 CTE makes an additional intramolecular interaction with the core DBD, through a loop, which stabilizes its contacts in the minor groove (5). CTEs can undergo conformational changes in response to DNA interaction. For example, CTE of RXR was reported to consist of a short α-helix in solution (6) but formed an extended loop conformation when bound to DNA (9,15). Conversely, the CTE of ERR-2 was unstructured in the absence of DNA and gained structure when bound to DNA (5,10).
The CTE is required for high-affinity DNA binding of many nuclear receptors (6,13,14,17–20), and for certain orphan receptors the CTE has been reported to play a role in specificity of target DNA by recognition of a preferred trinucleotide sequence flanking the half-site HRE (7,17,21,22). Recently, our crystal structure of a PR DBD–DNA complex showed for the first time that a steroid receptor CTE interacts with the minor groove flanking a HRE (23). Substitution mutations in two PR CTE residues (R637/K638) that insert into the minor groove reduced binding affinity for a palindromic PRE and eliminated binding to half-site PREs (23). Similar to orphan receptors, the CTE was shown by deletion mutagenesis to be essential for binding of PR and ER to half-site HREs (24,25). It has also been reported that acetylation of two lysine residues in the CTE of ER-α enhanced DNA binding and transcriptional activity (26). Thus the CTE of ER and PR participate directly in DNA binding and appear to be crucial for binding to weak half-site elements.
The chromosomal high-mobility group proteins B (HMGB) were first characterized as DNA architectural factors for various nuclear processes, including transcription, that require DNA manipulation through the ability of HMGB to bind to distorted DNA structures and to bend and partially unwind duplex DNA (27–37). We and others have shown that HMGB also functions to enhance the binding affinity of all classical steroid receptors to their cognate HREs in vitro, and consequently stimulate receptor transcriptional activity in transient cell transfection assays (24,25,38–43). HMGB-1 and closely related HMGB-2 are functionally interchangeable with respect to effects on steroid receptors; therefore, the generic term HMGB is used throughout unless a result was obtained with HMGB-1 or HMGB-2 specifically. In addition to binding the minor groove of DNA flanking PREs, the CTE is required for functional and physical interaction of steroid receptors with HMGB proteins. Studies with isolated DBDs of steroid receptors showed by deletion of the CTE, that this segment was required for protein interaction with HMGB and for the influence of HMGB on receptor DNA binding (24,25,42). Interestingly, HMGB has no effect on DNA binding or transcriptional activity of class II nuclear receptors (25). That protein interaction with the CTE is important for the stimulatory effect of HMGB on transcriptional activity of PR in intact cells was shown with PR/TR chimeric receptors. Swapping the CTE between PR and TR resulted in functional switching of HMGB's ability to enhance transcriptional activity from PR to TR (25). In addition to facilitating binding to consensus HREs, HMGB was shown to increase the affinity of ER for half-site EREs and for imperfect palindromic EREs, and to promote a greater fold increase for these intrinsically weak EREs than for consensus palindromic EREs (24,39). This may be of biological significance since few steroid receptor target genes contain consensus palindromic HREs. Further evidence that HMGB has a role in steroid receptor action in vivo comes from the phenotpye of HMGB-1 homozygous knockout mice in which thymocytes are resistant to glucocorticoid induced apoptosis (44). Also, an interaction between HMGB-1 and glucocorticoid receptor (GR) was detected by FRET in live cells (45). In addition to steroid receptors, HMGB can facilitate the activity of a number of other transcriptional activators (46–53), by enhancing their binding to cognate sequence specific target DNA. Interestingly, these studies also report a protein interaction with HMGB further indicating the general importance of protein–protein interaction in mediating the effect of HMGB on sequence-specific DNA binding by steroid receptors and other transcription factors. The mechanism for how this protein interaction affects DNA binding has not been elucidated.
Here, we further define the mechanism for how HMGB increases the binding affinity of PR for specific PRE DNA. Results in this article taken together with the recent structure of the PR CTE–DNA interaction (23), support the conclusion that HMGB increases binding affinity of PR for specific DNA primarily through protein–protein interaction with the CTE. The DNA bending property of HMGB was not required for enhancement of PR-DNA binding in vitro but was important for enhancement of transcription activity within intact cells. These data further suggest that manipulation of DNA by HMGB may be required either for access of PR to target PRE sites in the environment of chromatin or for other steps in the process of transcriptional activation and/or elongation.
MATERIALS AND METHODS
Materials
The preparation of the materials, including peptides, antibodies, recombinant baculovirus expression vectors, mammalian expression vectors and methods for protein expression and purification are described in the Supplementary Data. Wt and mutant HMGB-1 expression plasmids were obtained from Marco Bianchi (Milano, Italy) and have been previously described (45).
GST pull-down assays
Equal quantities of free GST or GST fusion proteins from bacterial lysates were incubated with 25 μl of GST-Sepharose resin for 1 h at 4°C. Resins were then washed three times (10 mM Tris pH 7.8, 50 mM NaCl, 10% glycerol, 2 mM MgCl2, 1 mM EDTA and 1 mM DTT), brought to a total volume of 250 μl in wash buffer and incubated with other proteins for 1 h at 4°C. Resins were then washed 4× in wash buffer, transferred to a clean tube and washed three more times. Bound proteins were eluted with SDS-sample buffer and detected by Western immunoblotting. In assays to determine the effects of DNA on protein interaction, 0.4 μM PR-A, PR DBD651 or PR DBD670 were incubated with 0.2 μM of a 26-mer PRE (5′GATATGAG AACAAACTGTTCTTAATC3′) or 20-mer nonspecific DNA (5′AGTTACTGAATTAC GCTCAT3′). In peptide competition assays, 1.5 μg of purified DBD-CTE670 or PR-B were incubated with peptide that was 50 to 5000 times the concentration of receptor. PR DBD651 measuring 0.4 μM was used in assays to determine the domains of HMGB-1 required for PR interaction. In assays to determine interaction with Mut HMGB-1 proteins versus Wt HMGB-1 proteins, PR DBD670 was added in increasing concentrations from 0.25 μM to 1.0 μM.
Electrophoretic mobility shift assay (EMSA)
EMSA was carried out as described previously (24,25,38). PR was preincubated for 30 min at 4°C (25 μl) in 10 mM Tris (pH 7.5), 50 mM NaCl, 5% glycerol, 2 mM MgCl2, 1 mM EDTA and 5 mM DTT in the presence of 0.1 μg poly(dA–dT) and 1 μg ovalbumin as a carrier protein. A PRE double-stranded oligonucleotide (5′GATCTTTGAGAACAAACTGTTCTTAAAACGAGGATC-3′) end labeled with 32P was added to reactions to a final concentration of 0.6 nM and incubated for 30 min on ice. Reactions were electrophoresed on 6% nondenaturing polyacrylamide gels (40 : 1 acrylamide/bisacrylamide ratio) in TAE buffer [0.02 M Tris–acetate (pH 8.0) and 0.5 mM EDTA] at 4°C. Gels were dried under vacuum at 80°C and the percentage of bound to free 32P labeled probe was determined with a series 400 Molecular Dynamics Phosphoimager. Experiments where Wt HMGB-1 and Mut HMGB-1 binding to DNA was examined, the same EMSA conditions were used excluding poly(dA–dT). Graphed binding curves were determined by averaging points from the minimum of three experiments (standard error shown) and fitting the averaged points to the equation:
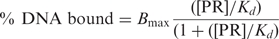
where Bmax is the maximum observed DNA binding, [PR] is the concentration of PR and Kd is the dissociation constant. The reported Kd was determined by fitting each binding experiment separately to the above equation then taking the mean from at least three experiments. Significance between Kds is reported as P from two-tailed type 2 t-tests.
Mammalian Cell Transfection
Transient transfections were performed by an adenovirus-mediated method as previously described with purified defective adenovirus particles covalently coupled to poly-l-lysine (54). Cos-1 cells were maintained in Dulbecco modified Eagle medium (DMEM, Gibco BRL) supplemented with 10% fetal bovine serum and were plated in six-well dishes (Falcon plates) at a density of 155 000 cells per well. Cells were transiently transfected with pCDNA1 plasmid containing HMGB-1(Wt) or (Mut) vector (or an empty pCDNA1 vector) along with a progestin-responsive PRE2-tk-LUC reporter, and an internal constitutive RSV-β-galactosidase reporter and a PR-B expression vector (24,38). The prepared adenovirus was mixed with plasmid DNA for 30 min at room temperature in HBS [20 mM HEPES (pH 7.8), 150 mM NaCl] followed by addition of poly-l-lysine at a 200-fold excess over plasmid DNA for another 30 min and added to cells at an MOI of 250. The cells were incubated for 24 h post-transfection and then treated with hormone (10 nM R5020) or EtOH for another 20 h at 37°C. At 48 h post-transfection, cell lysates were prepared and assayed for luciferase and β-galactosidase activities as described (24,38). Luciferase activity was determined and normalized to β-galactosidase as an internal control for transfection efficiency. Luciferase and β-galactosidase activities were analyzed on a Monolight 2010 luminometer.
Binding site selection
The binding site selection method was modified from Pollock and Treisman (55). Briefly, a polyclonal antiserum against PR DBD was used to immunoprecipitate complexes of PR DBD648 bound to DNA fragments containing a single inverted repeat PRE with a randomized 3N spacer and four randomized nucleotides on either side of the response element and flanked by fixed ends for PCR priming and subsequent subcloning. After the fifth cycle of immunopreciptation and amplification, the selected DNA was electrophoresed in the presence of 1 nM PR DBD-CTE648 on a 8% polyacrylamide gel (37.5 : 1 acrylamide:bis-acrylamide) for 90 min at 20 mA in TAE buffer (Tris–acetate-EDTA). The shifted DNA was visualized by Vistra Green-staining (Amersham Biosciences - now GE Healthcare, Invitrogen, USA), extracted via crush and soak methods, subcloned into the BamHI–EcoRI site of a pUC18 vector, and sequenced using standard methods (56). The sequences were aligned using CENSENSUS (57) and logos were created with Weblogo (58).
DNA circularization assays
The circularization assay was adapted from the previous studies (59). Assays were conducted with a 10-mer duplex DNA fragment with a cohesive two base over hang (5′-GCCTATTGAA-3′ and 5′-GCTTCAATAG-3′). Ligation reactions contained 2 μM HMG protein, 10 µM duplex DNA and 5000 cohesive end units T4 DNA ligase (NEB) in 100 μl ligase buffer (50 mM Tris–HCL pH 7.5, 10 mM MgCl2, 1 mM ATP, 10 mM DTT - NEB). Reactions were incubated at room temperature for 1 h before heat inactivation at 65°C for 10 min. Reactions were then digested with 100 U exonuclease III (NEB) at 37°C for 1 h before purification by PCI extraction. Samples were resuspended in 5% glycerol and load onto a 20 cm 8% (30:1 acrylamide:bis acrylamide; 1 × TBE) native gel. Gels were stained with Syber green (Invitrogen) and scanned with a series 400 Molecular Dynamics Phosphorimager.
RESULTS
The PR CTE is a specific protein-binding site for HMGB
We previously showed that the DBD CTE was the minimal fragment of PR able to mediate protein interaction with HMGB and to functionally respond to HMGB as detected by enhanced binding of PR to PRE DNA. Deletion of the CTE resulted in a loss of HMGB protein interaction and domain swapping of the CTE between class II receptors and PR resulted in a switch of functional response to HMGB (24,25). These results implicated a requirement of the CTE for physical interaction with HMGB and functional response to HMGB, but did not establish whether the CTE alone was sufficient for binding HMGB. As an initial experiment to address this question, we mapped HMGB interaction sites by GST pull-down assay with different fragments of PR. Ten different PR fragments containing 6× his tags were expressed in the baculovirus insect cell system and input and bound PR proteins were detected by Western immunoblot with an antibody to the 6× his tag (Figure 1). No specific binding was detected with the isolated amino terminal domains (NTD) of either PR-A or PR-B, or with the ligand-binding domain (LBD). Specific HMGB-1 interaction was detected with full-length PRs and only with PR fragments that contained the CTE with the exception of the DBD-hinge-LBD fragment that gave high nonspecific binding to free GST (Figure 1). This nonspecific interaction of the DBD-h-LBD does not preclude specific binding to the CTE. These results indicate that HMGB interaction requires the CTE and that HMGB does not bind specifically to other sites in the receptor.
Regions of PR and HMGB-1 required for protein interaction. PR constructs expressed in Sf9 cells with N-terminal 6× histidine tags were used in GST pull-down assay to detect HMGB-1 interaction. The position of the CTE in different PR constructs is indicated by a black line. Free GST or GST-HMGB-1 were immobilized on glutathione–Sepharose resins and incubated with Sf9 cell extracts containing his-tagged PR constructs. Bound PR was eluted from the resins and detected by western immunoblotting with a monoclonal mouse antibody (clone 1162/F6) to 6× his tag. Equal amounts of each PR protein were added as determined in advance by western immunoblot. Western blot lanes are: 10% input PR, free GST and GST-HMGB-1.
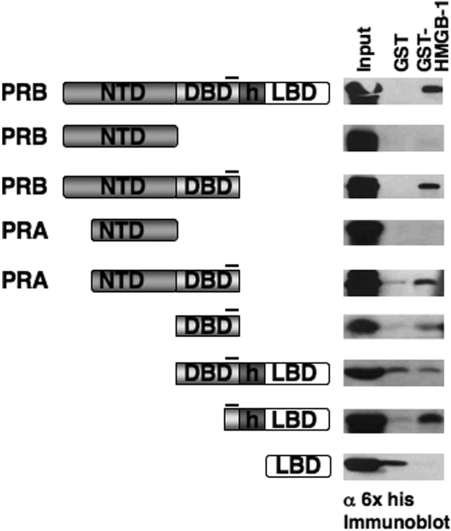
To determine whether the CTE in the absence of other potential intramolecular interactions is sufficient for interaction with HMGB, we analyzed the ability of synthetic peptides to compete for binding of HMGB-1 to PR (Figure 2A). Three peptides were used including a 19-mer peptide (CTE) that contains sequence corresponding to the PR CTE (aa 636–654), a scrambled peptide (sCTE) containing the same residues as the CTE except randomly scrambled to control for charge and amino acid composition, and an unrelated 19-mer peptide (URP) as a nonspecific control (60). These peptides were tested for their ability to compete for binding of HMGB-1 to PR in pull-down assays. Increasing concentrations of the CTE peptide inhibited binding of HMGB-1 to PR DBD-CTE670 as well as full-length PR. In contrast, the sCTE and URP peptides failed to inhibit binding (Figure 2B). These results, taken together with the PR domain mapping results (Figure 1), show that the CTE is both necessary and sufficient for binding to HMGB. The fact that the scrambled peptide was not able to inhibit protein interaction indicates that HMGB binding to the CTE is not simply a charge interaction but is mediated through a specific interaction surface.
The role of the CTE in HMGB-1 interaction. (A) Schematic of PR core DBD and CTE plus sequence of synthetic peptides corresponding to aa 636 to 654 in the CTE, scrambled sCTE peptide and a control unrelated peptide (URP). (B) CTE peptide specifically competed for binding of HMGB-1 to PR. Free GST and GST-HMGB-1 were immobilized to glutathione–Sepharose resins and incubated with purified PR DBD670 (0.5 µM/1.5 µg) or full-length PR-B (0.05 µM/1.5 µg) in the absence and presence of varying amounts of peptides. Peptides were added in molar excess over receptor; 50- to 250-fold for PR DBD670 and 500- to 5000-fold for PR-B. Resins were washed, eluted and bound receptor was detected by western blotting with antibody for PR DBD.
Both HMGB boxes are required for interaction with PR and stimulation of PR–DNA binding.
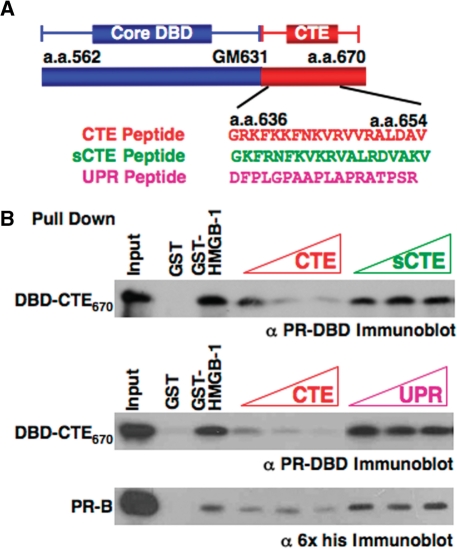
HMGB-1/-2 are expressed from separate genes and are composed of two homologous but nonidentical HMG boxes (box A and B), a basic region immediately C-terminal to each box, and an acidic C-terminal tail consisting of 30 or 20 Asp and Glu residues for HMGB-1 and HMGB-2, respectively (Figure 3A). Both HMG boxes have similar structure and bind to the minor groove of DNA in a sequence-independent manner but exhibit slightly different preferences for distorted DNA and ability to bend DNA (27,28,30,31,61–63). The minimal region of HMGB required for protein interaction with PR was defined using GST pull-down assays. Results showed that PR DBD–CTE651 bound as efficiently to an A and B di-box construct (termed Box AB or di-box) as it did to full-length HMGB-1, but the individual HMGB boxes bound much less efficiently to PR DBD-CTE651 (Figure 3B).
Regions HMGB-1 required for protein interaction. (A) Schematic of HMGB-1 constructs used in GST pull downs and EMSA. (B) Binding of HMGB-1 domains to PR DBD-CTE651. Whole-cell extracts prepared from bacterial cultures expressing GST, GST-box A, GST-box A with the basic linker (ABasic), GST-box B, GST-box B with basic linker (BBasic), GST-box A and box B (Box AB) or GST-HMGB-1 were immobilized on glutathione–Sepharose resin and incubated with 0.4 µM PR DBD–CTE651. Resins were washed, eluted and bound protein was detected by western blotting with antibody to PR DBD. (C) Enhancement of PR-DNA binding requires full-length HMGB-1 as shown by EMSA. Increasing amounts (0.5–7.0 μM) of HMG boxes were incubated with 32P labeled PRE in the presence of (2.0 nM) PR-A and compared to PR-A DNA binding alone and in the presence of (350 nM) full-length HMGB-2. Increasing concentrations of di-box (Box AB 0.5–2.0 μM) were incubated with 32P labeled PRE in the absence of PR-A to demonstrate Box AB's high-affinity nonspecific DNA binding. DNA binding was detected by EMSA as described in Materials and methods section.
Analysis of the effects of HMGB-1 domains on PR DNA binding is shown in Figure 3C. The individual HMG boxes, with or without the basic regions, had little or no ability to stimulate binding of PR-A to DNA even at a higher molar excess to PR than full-length HMGB-1 (200-fold) (Figure 3C). Analysis of the dibox was problematic since it acquired a substantial gain in affinity for general DNA as compared with full-length HMGB or individual box domains (Figure 3C) and bound efficiently to PRE–DNA in the absence of PR. Thus, at the amount required to stimulate PR-DNA binding, the dibox bound to the PRE–DNA and competed with PR binding (Figure 3C). This acquisition of DNA-binding affinity for the AB dibox in the absence of the acidic C-terminal tail has been previously described (64). At lower concentrations of the dibox that did not bind PRE, PR-DNA binding was not enhanced (data not shown). These data show that both HMG boxes are required for protein interaction with PR and that full-length HMGB is required for enhancement of PR-DNA binding.
Interaction between HMGB and the PR-CTE is disrupted by specific PRE DNA
In previous EMSA experiments, HMGB enhancement of PR-DNA binding affinity occurred without the presence of detectable HMGB in the final high-affinity receptor–DNA complex, indicating a stable ternary complex does not form with HMGB, PR and DNA (24,25,38–43). To further explore the mechanistic basis for this, we examined the influence of DNA on protein interaction between HMGB and PR DBD constructs containing varying lengths of the CTE (Figure 4A). In confirmation of previous results (25), HMGB-1 bound efficiently to the PR DBD containing CTE670 and CTE651, but did not bind to the more truncated CTE641, indicating that residues between 651 and 641 are required for interaction with HMGB-1 (Figure 4B). When excess specific PRE DNA was added to the binding reaction, HMGB interaction with PR DBD-CTE670 and PR DBD-CTE651 was prevented, whereas nonspecific DNA (NS) had no effect (Figure 4B). Similar results were observed for HMGB binding with full-length PR (A isoform); binding was inhibited by specific PRE DNA but not by addition of nonspecific DNA (Figure 4C). These results indicate that PR binding to PREs is not compatible with HMGB protein interaction and may explain why HMGB is not detected by EMSA in high-affinity PR–DNA complexes. Since the CTE is both a binding site for HMGB and participates in binding to DNA (minor groove flanking the PRE), these data also suggest the CTE uses the same or overlapping surfaces for protein and DNA binding.
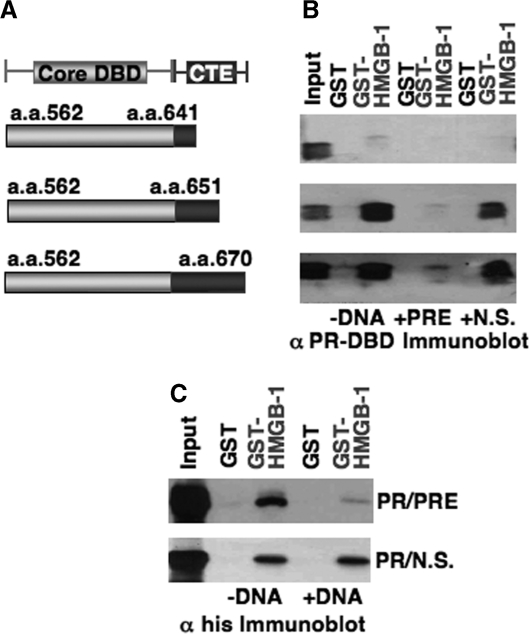
HMGB-1 protein interaction with CTE of PR is disrupted in the presence of PRE DNA. (A) Constructs of PR DBD with different lengths of CTE (aa 670, aa 651 and aa 641) were used in GST pull-down assays to detect HMGB-1 interaction. (B) Free GST or HMGB-1-GST fusion proteins were immobilized to glutathione–Sepharose resins as in Figure 1 and were incubated with PR DBD–CTE670, PR DBD–CTE651 or PR DBD–CTE641 in the absence (–DNA) or presence of PRE oligonucleotide (0.2 µM) or a nonspecific (N.S.) oligonucleotide DNA. Resins were washed, eluted and the bound protein was detected by western blotting with antibody for PR DBD. (C) GST pull-down assays with full-length PR were conducted as above in B and bound protein was detected by western blotting with a monoclonal antibody for PR.
A CTE peptide mimics HMGB enhancement of PR DNA binding
Since the CTE peptide inhibited interaction of HMGB with PR, we analyzed its influence on HMGB stimulation of PR–DNA binding with the expectation that it would compete for HMGB binding to the PR CTE and inhibit the ability of HMGB to stimulate PR–DNA binding. However, the CTE peptide enhanced binding of PR to the PRE DNA as detected by EMSA (Figure 5). The CTE peptide, in a dose dependent manner, increased the apparent DNA-binding affinity of the PR DBD-CTE670 by 5.8-fold (Figure 5A and Table 1). This effect was specific as the scrambled CTE and control peptides had minimal effect (Figure 5A and Table 1). The CTE peptide had only a minimal effect (1.3-fold) on DNA binding by the truncated PR DBD-CTE 641 construct (Figure 5B and Table 1). It should be noted that for the truncated DBD-CTE641, the DNA-binding affinity in the absence of the CTE peptide increased by ~2-fold as compared to PR DBD-CTE670 (Table 1). This is consistent with our previous observation that the CTE has a suppressive effect on ER and PR DNA-binding activity (24,25). However, the stimulatory effect of the CTE peptide and HMGB on DNA-binding affinity of PR is larger (8–9-fold) than the 2-fold reduction observed by truncation of the CTE to aa 641. The ability of the CTE peptide to mimic the effect of HMGB on PR–DNA binding in a manner dependent on the presence of CTE in the PR construct is consistent with the hypothesis that the flexible CTE in the absence of DNA makes contact with other regions of PR that represses DNA-binding activity. Such an intramolecular interaction could be disrupted either by HMGB binding to the CTE or by competition with an external CTE peptide that binds with the core DBD and disrupts this intramolecular interaction.
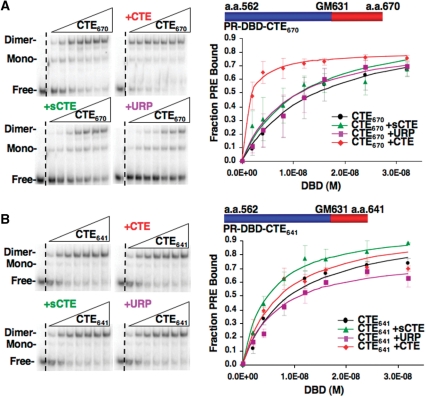
The role of the CTE in PR-DNA binding. (A and B) CTE peptide enhances PR–DNA binding in a manner dependent on the presence of CTE in the receptor. Increasing amounts (2–32 nM) of (A) PR DBD-CTE670 or (B) PR DBD–CTE641 were incubated with 32P labeled PRE in the presence or absence of CTE, sCTE and URP peptide (0.75 µM), and DNA binding was detected by EMSA as described in Materials and methods section. Binding was graphed as fraction PRE bound and represent average values ± standard error of the mean (SEM) from a minimum of three independent experiments.
Table 1.
Fold enhancement of PR DBD by CTE peptides
| (-) | URP | sCTE | CTE | |
|---|---|---|---|---|
| DBD-CTE670 Kd(M) ± SEM [Fold effect/P(T ≤ t)] | 1.69E−08 ± 6.06E−09 | 1.38E−08 ± 4.56E−09 (1.2/0.55) | 1.12E−08 ± 2.66E−09 (1.5/0.29) | 2.90E−09 ± 1.16E−09 (5.8/0.14) |
| DBD-CTE641 Kd(M) ± SEM [Fold effect/P(T ≤ t)] | 9.25E−09 ± 7.24E−10 | 1.06E−08 ± 3.62E−10 (NA/0.29) | 4.31E−09 ± 2.37E−10 (2.1/0.01) | 7.34E−09 ± 1.48−10 (1.3/0.46) |
HMGB influences the preference of PR for A/T sequences flanking the PRE
Our previously observed interactions of the PR CTE with the minor groove flanking the PRE (23), taken together with results here, raised the question of whether HMGB influences receptor recognition of sequences flanking the hexa-nucleotide PRE. To address this question, we conducted a binding-site selection experiment with DNA containing a fixed inverted repeat hexanucleotide consensus PRE with a randomized 3N spacer and four randomized nucleotides flanking either side of the PREs (N4AGAACAN3TGTTCTN4). In the absence of HMGB, PR DBD-CTE648, showed a slight strand bias at positions −2 and +7 in the PRE and nonrandom distribution of bases in the 3N spacer and flanking DNA as detected by CENSENSUS (57) and illustrated by Weblogo (58), (Figure 6). These aligned sequences had a preference for pyrimidines at positions −1 and 0 in the 3N spacer. However, in the presence of HMGB-1, PR DBD-CTE648 had a preference for pryimidines only at position 0, and for A/T residues at positions +8, +9 and +11 in the flanking sequence that was not observed with PR DBD- CTE648 alone (Figure 6). HBGB-1 also promoted a slight strand bias within the fixed PRE at positions +3 and +7 (Figure 6). These results indicate that HMGB asymmetrically influences the flanking PRE sequence recognized by PR and thus may alter the specificity of PR target DNA.
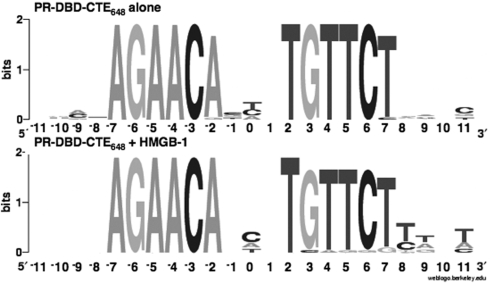
HMGB-1 influences PRE flanking DNA sequence recognized by PR. Shown in the Sequence logo format (Weblogo) are the sequences selected by PR DBD–CTE648 alone or in the presence of HMGB-1 (+HMGB-1) as aligned by CONSENSUS. The sequence positions of the inverted repeat PREs are shown on the x-axis and the information content of the alignment for each position is shown on the y-axis.
DNA architectural properties of HMGB-1 are not required for enhancement of PR–DNA binding
DNA-binding characteristics of box A and box B of HMGB are attributed in part to three minor groove intercalating residues that are conserved among HMGB family members (27,65). Box A contains one intercalating residue (Phe37) and box B contains two intercalating residues (Phe102 and Ile121) (Figure 7A). The basic regions C-terminal to the HMG domains have also been shown to be involved in DNA interactions and are required for nuclear accumulation (66,67) (Figure 7A). The role of the DNA-binding properties of HMGB in facilitating PR interaction with PRE was investigated using a previously described mutant HMGB-1 (Mut HMGB-1) which contains alanine substitutions of the three key intercalating residues (45). To assess DNA-binding properties of Mut HMGB-1, EMSAs were performed with varying concentrations of Wt HMGB-1 and Mut HMGB-1 using PRE DNA as a target. Because HMGB-1 binds to DNA in a nonsequence-specific manner, no carrier DNA was added to the reactions to permit detection of low-affinity binding. Wt HMGB-1 bound the PRE DNA and had ~5–6-fold higher binding affinity (Kd = 2.03E−07) than Mut HMGB-1 (Kd = 1.43E−06). Despite the lower binding affinity, the Mut HMGB-1 exhibited a higher maximal DNA binding (15% versus 5% of input DNA) (Figure 7B and Table 2). Wt HMGB-1 and Mut HMGB-1 complexes also exhibited distinct mobilities indicating differences in how they distort DNA structure (Figure 7B). Ligase-mediated circularization assays were preformed to determine the ability of the wild-type and mutant HMGB-1 to bend DNA (59) (Figure 7C). It has been previously established that ligation of short linear DNA fragments in the presence of HMG proteins produces DNA circles with sizes that relate to the degree to which HMG is able to bend the DNA (63,65,68,69). A 10-mer DNA fragment was ligated in the presence of HMGD, Wt HMGB-1, Mut HMGB-1 or no protein. Drosophila HMGD was used as a positive control since its DNA-bending properties have been extensively characterized (59,70). Wild-type HMGB-1 was able to efficiently produce circles of 80 bp in size and larger by increments of 10 bp with activity similar to HMGD (Figure 7C). In contrast, the Mut HMGB-1 demonstrated almost a total loss of DNA bending producing only a small proportion of circles >150 bp (Figure 7C). Therefore, Mut HMG-1 binds to DNA with slightly lower affinity compared to Wt-HMGB-1, but is essentially unable to bend DNA.
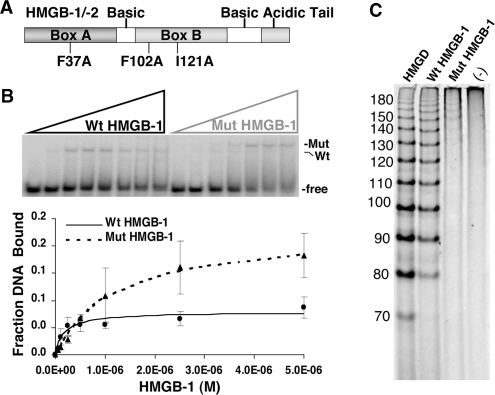
Functional effect of mutations in the DNA intercalating residues of HMGB-1. (A) Schematic of HMGB-1 domains. Mut-HMGB-1 contains alanine substitutions for the residues that intercalate in the DNA (Phe37, Phe102 and Ile121) as described previously (45,62). (B) Binding of wild-type (Wt) and mutant (Mut) HMGB-1 with PRE DNA as detected by EMSA. Increasing concentrations of WT HMGB-1 or Mut HMGB-1 proteins (0–5 µM) were incubated with a single concentration of [32P]-labeled PRE (0.6 nM) under conditions in the absence of competitor DNA that permit the detection of nonsequence-specific DNA binding. The distinct mobilities of Wt-HMGB-1 and Mut HGMB-1 complexes are indicated by the arrows. Free and shifted DNA bands were quantitated and graphed as a fraction of bound DNA. (C) DNA-bending properties of HMGB-1 Wt and Mut proteins as analyzed by circularization assay. HMGD, HMGB-1 Wt, HMGB-1 Mut (2 μM) or no protein was incubated with 10-mer linear duplex DNA in the presence of DNA ligase. Reactions were digested with exocnuclease III to eliminate linear DNA fragments. HMGD as a positive control was used to generate a ladder of known DNA circle sizes. Formation of DNA circles was detected by EMSA and cyber green staining (Invitrogen) as described in Materials and methods section.
Table 2.
Binding affinity of Wt and Mut HMGB-1
| Wt HMGB-1 Kd (M) | 2.03E−06 ± 5.79E−07 |
| Mut HMGB-1 Kd(M) | 1.43E−06 ± 5.52E−07 |
| P(T ≤ t) | 0.36 |
Mutant and wild-type HMGB-1 were also compared for their ability to make protein contacts with PR DBD-CTE670 and to facilitate PR–DNA binding. In GST pull-down assays, Wt HMGB-1 and Mut HMGB-1 interacted equally with PR (Figure 8A). Thus, the HMGB-1 intercalating residues are not required for protein interaction with PR. Both Wt HMGB-1 and Mut HMGB-1 had similar effects on PR–DNA binding stimulating an increase in apparent binding affinity of 7–8-fold from a Kd of 1.18E−07 with receptor alone to a Kd of 1.60E−08 and 1.48E−08 with Wt HMGB and Mut HMGB-1, respectively (Figure 8B and Table 3). These results show that the DNA intercalating residues of HMGB-1 are not required in vitro for protein interaction with PR or for stimulation of PR–DNA binding.
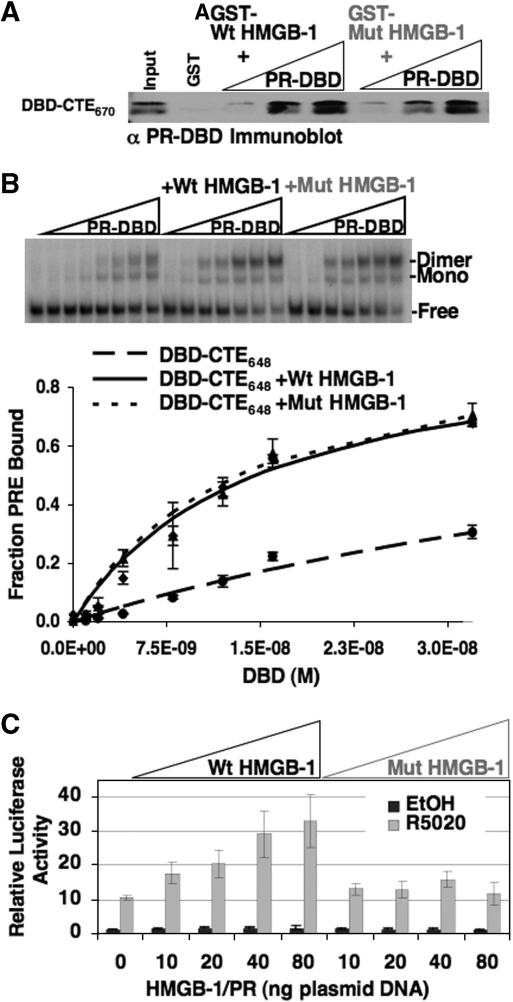
Mutations in DNA intercalating residues of HMGB-1 did not influence enhancement of PR-DNA binding in vitro but reduced the effect of HMGB-1 on PR transcriptional activity in cells. (A) Protein interaction of HMGB-1 with PR DBD was not affected by mutations in the intercalating residues of HMGB-1. Free GST, GST-Wt HMGB-1 or GST-Mut HMGB-1 were immobilized to glutathione–Sepharose resins and incubated with increasing concentrations of purified PR DBD–CTE670 (0.25 µM to 1.0 µM). Resins were washed, eluted and bound protein was detected by western blotting with antibody for PR DBD (input is 10%). (B) Effect of Wt HMGB-1 and Mut HMGB-1 on binding of PR to PRE DNA as detected by EMSA. Increasing concentrations of purified PR DBD–CTE648 (0–30 nM) were incubated with a single concentration of [32P]-labeled PRE (0.6 nM). The free and protein bound PRE complexes were separated by native gel electrophoresis, quantitated, and graphed as fraction of bound PRE–DNA. (C) Mut HMGB-1 had reduced ability to enhance transcriptional activity of PR in cells. Cos-1 cells were cotransfected with PRE2-tk-LUC reporter, PR-B, Wt HMGB-1 or Mut HMGB-1. Cells were treated with 10 nM R5020 (synthetic progesterone analog) or an equal volume of EtOH for 20 h. Luciferase activity was determined and normalized to internal control β-galactosidase activity. Normalized luciferase activity with vehicle-treated cells was set to 1.0 and all other treated groups were calculated as fold >1.0. Error bars indicate standard mean of the error (n = 5) for doses 0–40 and n = 3 for dose 80.
Table 3.
Fold enhancement of PR DBD by Wt and Mut HMGB-1
| (-) | Wt HMGB-1 | Mut HMGB-1 | |
|---|---|---|---|
| DBD648 Kd(M) ± SEM [Fold effect/P(T ≤ t)] | 1.18E−07 ± 2.35−08 | 1.60E−08 ± 7.38E−10 (7.4/0.02) | 1.48E−08 ± 3.09−10 (8.0/0.02) |
Mut HMGB-1 was also compared with Wt HMGB-1 in transient transfection assays for their ability to stimulate transcription activity of PR (Figure 8C). Cos cells were cotransfected with a PRE-luciferase (LUC) reporter gene, PR-B, Wt HMGB-1, Mut HMGB-1 or an empty vector. Cells were treated with R5020, a synthetic progesterone, or vehicle (EtOH). Under conditions where ectopically expressed wild-type HMGB-1 gave a significant enhancement of progesterone-dependent PR-mediated transactivation of PRE-Luc, the Mut HMGB-1 had minimal effect (Figure 8C). These results show that even though intercalating residues of HMGB-1 were not required for PR interaction or stimulation of PR–DNA binding they were important for stimulation of PR transcription activity in cells.
DISCUSSION
Results of the experiments presented in this paper provide new insights into the mechanism by which HMGB proteins enhance the activity of PR. First, the CTE of PR contains a motif that is recognized by HMGB and this protein interaction was found to be essential for HMGB enhancement of PR–DNA binding in vitro. Second, specific PRE DNA interfered with HMGB–CTE interaction suggesting that CTE binding with the minor groove of DNA flanking the PREs and with HMGB are mutually exclusive. This may explain previous results from our lab and others on the inability to observe a stable ternary complex of steroid receptor/HMGB/DNA. Third, a CTE peptide mimicked HMGB in vitro by enhancing PR–DNA binding in a manner dependent on the presence of the CTE in the PR construct. This taken together with previous results that truncation of the CTE increased the affinity of PR for PRE DNA, suggests that HMGB interaction relieves a repressive effect of the CTE on PR DNA binding. Fourth, the presence of HMGB in a DNA-binding site selection assay resulted in PR exhibiting a preference for A/T nucleotides flanking the PRE indicating that HMGB influences target gene specificity. Finally, the DNA binding and bending activities of HMGB-1 were not required for protein interaction with PR or stimulation of specific PR–DNA binding in vitro but were required for HMGB-1 enhancement of PR-mediated gene transcription in intact cells. These data suggest that HMGB-1 affects more than PR–DNA binding and may enhance other steps in the process of PR- mediated activation of gene transcription.
From these results we propose a working hypothesis for how HMGB stimulates PR–DNA binding in vitro and PR-mediated transcriptional activation in intact cells (Figure 9). HMGB interacts with the flexible CTE prior to PR–DNA binding, resulting in disruption of an intramolecular interaction between the CTE and core DBD that is repressive for DNA binding. This HMGB interaction also promotes binding of the CTE to the minor groove of DNA immediately flanking the PRE. The CTE–DNA interface increases the affinity of the PR DNA complex and has a preference for A/T nucleotides suggesting that HMGB also alters target gene specificity. HMGB interaction with PR is transient and is dissociated upon binding of the CTE to DNA. In the intact cell HMGB recruited by PR to target gene sites may have other roles that require DNA-binding and -bending activities of HMGB such as facilitating access of PR to target genes in chromatin and other steps in the process of activation of gene transcription.
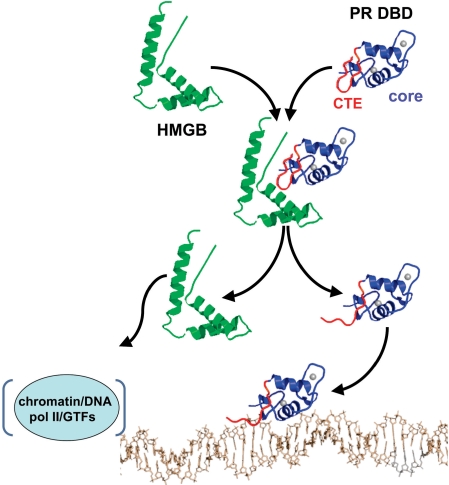
Proposed mechanism of action of HMGB. Prior to binding DNA, HMGB (green) interacts with the CTE (red) of PR (core DBD in blue) and disrupts a repressive intra-molecular interaction between the CTE and core DBD (only single domain models are shown for simplicity). HMGB interaction also promotes CTE binding to the DNA minor groove flanking the PRE. HMGB binding to PR is transient and dissociates upon stable binding of the CTE to DNA. Dissociated HMGB interacts with DNA/chromatin in the vicinity of the promoter to affect RNA pol II and general transcription factors (GTFs) in down stream steps in the process of steroid receptor-mediated gene transcription.
HMGB influences the DNA-binding specificity of PR
In a binding-site selection assay, HMGB-1 influenced sequences flanking the PRE that were recognized by the PR DBD–CTE. In the presence of HMGB-1, an increased occurrence of A/T-rich sequences at nucleotide positions +8, +9 and +11 adjacent to the 3′ PRE hexanucleotide (Figure 6) was observed. Interestingly, the PR DBD dimer binds asymmetrically to DNA, as seen in the crystal structure of the PR DBD–DNA complex, due to the 3N spacer (23). The DBD subunit in the crystal structure that had weaker contacts was bound to the 3′PRE hexanucleotide and this correlated with the selection of A/T-rich sequences by HMGB in the 3′ flanking sequence (Figure 6). These data indicate that HMGB-1 either altered the interaction of the CTE with the flanking DNA or transiently bound the CTE to further stabilize weak or suboptimal PR–DNA interactions through the flanking DNA. Precedence for this role of the CTE comes from certain orphan receptors that bind to half-site HREs, where CTE interactions with the minor groove extend the DNA-binding site through a preference of the CTE for a flanking TCA(RevErb) or AAA (NGFIB) trinucleotide. A preference of PR for A/T-rich flanking sequences in the binding-site selection assay with HMGB is consistent with a well-characterized preference of HMGB to bind A/T-rich DNA sequences in general (68,70,71). The influence of HMGB on steroid receptor specificity for DNA has been observed previously for ER (39). The combined effect of the CTE and HMGB to maximize receptor interaction with weak half-site HREs may be of biological significance since few steroid receptor target genes contain consensus HREs. The majority have divergent imperfect HREs or multiple half-site HREs suggesting that auxiliary cofactors are required for receptors to select many of their target genes in vivo. HMGB is an abundant cellular protein that may play such a role and expand the diversity of genes recognized by steroid receptors beyond those with optimal palindromic HREs.
HMGB induced de-repression of PR DNA binding is mediated by the CTE
Previous reports showed that truncation of the CTE in the context of PR DBD and ER-DBD constructs resulted in an increased DNA-binding affinity (24,25). However, addition of HMGB increased DNA-binding affinity more than that observed by truncation of CTE. This information combined with the result that the CTE peptide increased PR–DNA binding affinity in a manner dependent on the presence of the CTE in the PR DBD polypeptide, is consistent with a ‘relief from repression’ mechanism that involves HMGB binding to the CTE (Figure 5). It has previously been shown in the structure of the orphan receptor ERR2, that residues in the CTE interact with a hydrophobic patch on the core DBD through looping of a region of the CTE beyond the residues that insert in the minor groove flanking the HRE. (5). The effect of the CTE intramolecular interaction with the core DBD in ERR-2 has not been directly tested. Interestingly, our PR DBD–CTE648 crystal structure showed a similar hydrophobic patch in the core DBD that could potentially mediate an intramolecular interaction with the CTE (23). However, the PR DBD–CTE648 structure did not have enough of the CTE sequence present to detect such an interaction if it exists. Since the CTE is known to be a flexible region in dynamic equilibrium between different conformations, these data suggest that the CTE peptide enhanced PR–DNA binding by competing for an intramolecular interaction between the CTE and core DBD. Further structural studies with a longer CTE and functional mutagenesis will be needed to confirm this proposed mechanism.
Direct interaction of HMGB and the CTE is important for enhancement of PR–DNA affinity and activity
A common finding with transcription factors (including steroid receptors) that exhibit increased DNA-binding affinity in response to HMGB proteins is that HMGB is not (at least in vitro) a stable component of the high-affinity DNA complex (38–53). Thus the effect of HMGB is mediated through transient interactions, either by altering the conformational equilibria of receptor-DNA interactions, or the conformation of the DNA to improve it as a target for receptor interaction. Since HMGB proteins bind and distort DNA it was logical to anticipate that HMGB acts by altering the orientation or accessibility of the DNA. To examine the role of DNA architectural properties, we employed mutations of DNA intercalating hydrophobic residues of HMGB-1. These substitutions were previously reported to reduce binding to structured and linear nonspecific DNA and to reduce DNA-bending activities of the individual HMG boxes (57,65,66,72). Our results confirm the reported functional consequence of these substitutions within the context of the full-length protein. The mutations did not abolish HMGB DNA binding, rather they reduced affinity for nonspecific linear DNA. However, DNA-bending activity of the mutant HMGB1 was completely lost (Figure 7B and C). These substitutions had no effect on protein interaction with PR or the ability of HMGB-1 to stimulate DNA-binding affinity of PR in vitro (Figure 8A and B). Based on these results we conclude that HMGB, at least under these conditions in vitro, does not increase PR–DNA-binding affinity by manipulating structure of the DNA template. HMGB acts primarily through protein–protein interaction with PR to facilitate interaction of the CTE with DNA sequences flanking the PRE.
In contrast to the lack of effect on PR–DNA binding in vitro, these intercalation site mutations markedly reduced HMGB-1 stimulation of PR transcriptional activity in cell transfection experiments (Figure 8C). An analogous observation was made by Agresti et al. (45), where interaction between GR and HMGB-1 observed in living cells by FRET was dependent on both an intact DBD of GR and the intercalating residues of HMGB-1. The authors concluded that both GR and HMGB-1 must be bound to DNA in order to interact with each other. Thus, one possible explanation for the different effects of HMGB mutations observed in vitro and in intact cells, is that the DNA intercalation and bending properties of HMGB are required to enable access of PR to its DNA-binding sites in the environment of chromatin. Indeed, HMGB has been shown previously to act in a chromatin remodeling capacity (73). Alternatively, HMGB-1 may have additional effects on the process of receptor-mediated transcription downstream of PR–DNA binding. HMGB has been reported to interact with TBP at the initiation complex (74) and to be a component of an elongation complex associated with RNA polymerase in vitro (75). Since chromatin remodeling and elongation likely require unwinding of DNA, the DNA intercalating properties of HMGB could be essential to these processes. Thus transient interaction of HMGB with receptors could have the dual role of facilitating receptor–DNA binding and localizing HMGB at the promoter for interaction with general transcription factors or elongation complexes. Further studies will be needed to determine whether HMGB affects steps other than receptor–DNA binding in the process of steroid receptor-mediated transcriptional regulation.
ACKNOWLEDGEMENTS
This work was supported by Public Health Services Grant NIH CA46938 (DPE). Funding to pay the Open Access publication charges for this article was provided by PHS NIH grant CA46938 (DPE).
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkn249
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/36/11/3655/17422577/gkn249.pdf
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/abstract/36/11/3655
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/content/full/36/11/3655
Free to read at nar.oxfordjournals.org
http://nar.oxfordjournals.org/cgi/reprint/36/11/3655.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
CRABS CLAW Acts as a Bifunctional Transcription Factor in Flower Development.
Front Plant Sci, 9:835, 20 Jun 2018
Cited by: 37 articles | PMID: 29973943 | PMCID: PMC6019494
Repeated abortion in adulthood induces cognition impairment in aged mice.
Sci Rep, 8(1):11396, 30 Jul 2018
Cited by: 1 article | PMID: 30061639 | PMCID: PMC6065382
Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast.
Nucleic Acids Res, 45(14):8378-8391, 01 Aug 2017
Cited by: 15 articles | PMID: 28637278 | PMCID: PMC5737332
Kilobase Pair Chromatin Fiber Contacts Promoted by Living-System-Like DNA Linker Length Distributions and Nucleosome Depletion.
J Phys Chem B, 121(15):3882-3894, 31 Mar 2017
Cited by: 22 articles | PMID: 28299939 | PMCID: PMC6203935
The DNA chaperone HMGB1 potentiates the transcriptional activity of Rel1A in the mosquito Aedes aegypti.
Insect Biochem Mol Biol, 80:32-41, 17 Nov 2016
Cited by: 8 articles | PMID: 27867076
Go to all (21) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The role of the C-terminal extension (CTE) of the estrogen receptor alpha and beta DNA binding domain in DNA binding and interaction with HMGB.
J Biol Chem, 279(15):14763-14771, 21 Jan 2004
Cited by: 35 articles | PMID: 14739282
A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain.
J Biol Chem, 284(36):24415-24424, 24 Jun 2009
Cited by: 25 articles | PMID: 19553667 | PMCID: PMC2782034
Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements.
Mol Endocrinol, 20(12):3042-3052, 24 Aug 2006
Cited by: 47 articles | PMID: 16931575 | PMCID: PMC2532839
Structural and functional analysis of domains of the progesterone receptor.
Mol Cell Endocrinol, 348(2):418-429, 22 Jul 2011
Cited by: 51 articles | PMID: 21803119 | PMCID: PMC4437577
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA046938
Grant ID: CA46938
NIGMS NIH HHS (5)
Grant ID: R01 GM059456-05
Grant ID: R01 GM059456-03
Grant ID: R01 GM059456-04
Grant ID: R01 GM059456
Grant ID: R01 GM059456-02




