Abstract
Free full text

Quantifying environmental adaptation of metabolic pathways in metagenomics
Abstract
Recently, approaches have been developed to sample the genetic content of heterogeneous environments (metagenomics). However, by what means these sequences link distinct environmental conditions with specific biological processes is not well understood. Thus, a major challenge is how the usage of particular pathways and subnetworks reflects the adaptation of microbial communities across environments and habitats—i.e., how network dynamics relates to environmental features. Previous research has treated environments as discrete, somewhat simplified classes (e.g., terrestrial vs. marine), and searched for obvious metabolic differences among them (i.e., treating the analysis as a typical classification problem). However, environmental differences result from combinations of many factors, which often vary only slightly. Therefore, we introduce an approach that employs correlation and regression to relate multiple, continuously varying factors defining an environment to the extent of particular microbial pathways present in a geographic site. Moreover, rather than looking only at individual correlations (one-to-one), we adapted canonical correlation analysis and related techniques to define an ensemble of weighted pathways that maximally covaries with a combination of environmental variables (many-to-many), which we term a metabolic footprint. Applied to available aquatic datasets, we identified footprints predictive of their environment that can potentially be used as biosensors. For example, we show a strong multivariate correlation between the energy-conversion strategies of a community and multiple environmental gradients (e.g., temperature). Moreover, we identified covariation in amino acid transport and cofactor synthesis, suggesting that limiting amounts of cofactor can (partially) explain increased import of amino acids in nutrient-limited conditions.
Microbes function as highly interdependent communities. Fundamental to the maintenance of the energy balance of the ecosystem, the recycling of nutrients, and the neutralization and degradation of toxins and other detritus (1), microbial community processes are intimately intertwined with ecosystem functioning. Thus, it is critical to understand the complex interplay between the influence of the environment on microbial communities, and, in turn, the microbes' reshaping of their environment.
Until recently, the tools to systematically study global community function and environment at the molecular level were not available, because complex microbial communities are generally not amenable to laboratory study (2). The recent advent of direct sequencing of environmental samples (i.e., metagenomics) has allowed the first large-scale insights into the function of these complex microbial communities.
Comparative metagenomics approaches have revealed significant variation in sequence composition (3), genome size (4), evolutionary rates (5), and metabolic capabilities (6–8) among qualitatively dissimilar environments (e.g., terrestrial vs. marine), providing evidence for genomic adaptations. Further, variation in specific community biological processes have been shown for different water column zones at a single geographic site (9), different climatic regions in the ocean (10), and, more recently, among 9 ecosystems (7).
The wealth of information generated from these studies emphasizes the importance of investigating relative differences in biological processes among qualitatively different environments. However, to date, none of them have directly incorporated multiple, specific measurements of the environment. By treating the environment explicitly as a set of complex, continuous features, rather than relying on an implicit subjective classification, one can build models to determine how a diverse array of biochemical activities, and particularly metabolic versatility, reflect sets of or specific environmental differences.
Providing an ideal dataset for exploring these environmental–biochemical links, the Global Ocean Survey (GOS) collected quantitative environmental features and metagenomic sequences from >40 different aquatic sites (10). Here, we used GOS data to investigate and develop multivariate approaches to systematically relate metabolic pathway usage directly to quantitative environmental differences. These approaches allowed us to address multiple relationships simultaneously, as well as to relate specific environmental features to metabolic processes at different levels of resolution, including 14 broad functional categories, 111 pathways, 141 modules (sections of pathways), 191 operons, and 15,554 orthologous groups (OGs). By identifying environmentally-dependent pathways involved in energy conversion, amino acid metabolism, and cofactor synthesis, among others, we were able to define metabolic footprints of distinct environments. Our study provides an analytical framework for uncovering ways, in which microbes adapt to (and perhaps even) how they change their environment.
Results
Quantitative Approach for Footprint Detection.
We mapped 37 size-filtered GOS sites (Table S1) to their respective environmental and metabolic features at several levels of complexity (pathways, modules, and operons; see Fig. 1 A and B). These data can naturally be represented as matrices, where the rows are geographic sites and the columns are either environmental or metabolic features. We interrelated these matrices to examine how pathway usage across different sites is related to environmental parameters. The simplest and most direct approaches for performing such operations are correlation and regression (for comparisons with other types of methods see Fig. S1 and Table S2). Thus, we examined the first order relationships by computing the pairwise correlation between each metabolic pathway and each environmental feature (e.g., photosynthesis and temperature). Note that for clarity, we use the word pathway to refer to the usage of the pathway, as in photosynthesis as opposed to usage of photosynthesis, in the remainder of the text. This analysis revealed a number of significant correlations (environmentally-dependent pathways; see Table S3). Such pathways were used to build linear models (LM) of each environmental feature (see Table S4). Although these models performed well in predicting single environmental features (Fig. 2), there are limitations to viewing each environmental measurement in isolation, because there are hidden dependencies among the environmental features.
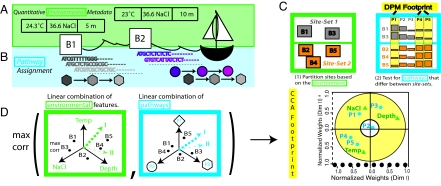
Schematic representation of approach. The large squares labeled B1, B2, etc. represent the geographic sites (buckets). Each bucket has sequence and environmental feature data associated with it. (A) Mapping quantitative environmental features [salinity (ppt), sample depth (position in water column from which the sample was collected), water column depth (measured from surface to floor), and chlorophyll]. (B) Metagenome-derived metabolism at different levels of resolution (see Materials and Methods). Reads are color-coded according to their corresponding pathway elements (shapes). Different pathways are represented by different shapes (square, circle, etc.). All of the instances of a particular pathway are summed and normalized to compute the pathway score. (C) Schematic representation of DPM (see details in text). (D) Schematic representation of CCA (see details in text).
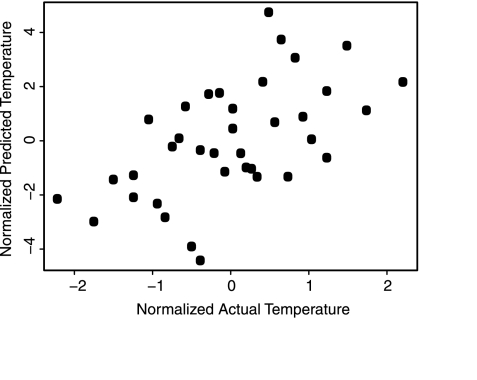
Predicting specific environmental parameters from subsets of metabolic pathways. Linear model for temperature built from subsets of highly correlated pathways, including N-acetylglucosamine biosynthesis, many components of amino acid metabolism, and fatty acid biosynthesis (for full list and coefficients, see Table S4). Axes are normalized actual and predicted temperature for x and y, respectively.
To discover the complex, higher order interactions between and within environmental features and metabolism, we used a second complementary approach, regularized canonical correlation analysis (CCA). CCA has 2 primary functions: (i) to determine whether a global relationship between 2 types of features (here, environmental and metabolic) exists; and (ii) to calculate the relative contribution of each feature to the global relationship (e.g., temperature or photosynthesis), by weighting both sets of features simultaneously (11). In brief, CCA computes a linear combination for each feature set and simultaneously attempts to maximize the correlation between the 2 feature vectors (Fig. 1D). Thus, CCA is able to simultaneously assess relationships both between and among the environmental features and metabolic pathways. Because the sites are quite similar, we developed a more robust but less sensitive method called discriminative partition matching (DPM). DPM first partitions the sites into site sets on the basis of their environmental parameters, then tests which pathways give the greatest discriminatory power among the site sets (Fig. 1C).
Footprint Characteristics.
The goal of DPM and CCA is to simultaneously explore the relationship between metabolism and the quantitative environmental parameters by identifying environmentally-dependent or covarying metabolic pathways (footprints). The main difference between DPM and CCA is that DPM identifies those pathways that discriminate the best between site sets, but when defining the site sets, all of the environmental variables are considered equally important. Thus, although robust to noise, DPM is more coarse-grained, and, at this resolution, the individual differences among sites and their relationship to the environment can be lost. In contrast, CCA can highlight these individual differences by weighting each environmental feature and each metabolic pathway independent of any partitioning, making it both more sensitive but also more susceptible to noise (Fig. 1D).
DPM Footprint.
Applying DPM, the sites were partitioned into 2 different site sets that can loosely be classified as open ocean and coastal. We found that the distribution of those clusters of orthologous groups of proteins (COGs) and Kyoto Encyclopedia of Genes and Genomes (KEGG) maps annotated as having a role in metabolism was significantly different between site sets (P < 9 × 10−3 and 4 × 10−14, respectively); however, no statistically significant difference was found for control matrices that were composed of translational/transcriptional machinery (see SI Materials and Methods).
Also, we find 10 KEGG maps, 24 modules, 61 operons, and 98 gene families were significantly different [false discovery rate (FDR)-corrected q < 0.05] between the 2 site sets. These pathways together form the DPM footprints (Table S5). By examining the broader trends of these footprint pathways, we found that secondary metabolite biosynthesis, lipid transport and metabolism, amino acid metabolism, and energy production and conversion were significantly different between site sets. Finally, we showed that the cluster similarity between the environment-based site partitioning and metabolic footprint-based site partitioning was quite high (normalized mutual information, 0.46; rand index, 0.76; P < 0.001), suggesting that footprints have predictive power in recapturing features of the environment based purely on pathways identified as significant in DPM.
CCA Footprint.
Next, we applied regularized CCA to measure the strength of the metabolic pathway's covariation with environmental features. We identified 22 KEGG maps, 53 modules, and 35 operons as being environmentally dependent (absolute value of structural correlations >0.3; see Fig. 3). These pathways form footprints that can be investigated for subtler environment-based changes in metabolic capabilities (Table S6). In this manner, we identified diverse functional processes that covaried significantly with the environment, including xenobiotic degradation, energy conversion, lipid metabolism, and amino acid metabolism.
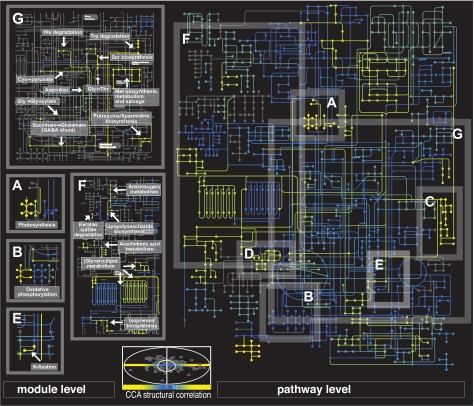
Metabolic map of structural correlations at 2 resolutions. Central panel is a plot of the environmental features (triangles) and pathways (circles), where the x axis and y axis are the structural correlation coefficients (normalized weights) derived from CCA for the first and second dimension, respectively (see Fig. 1D). The remainder of the figure indicates the strength of both the environmental covariation of the pathways (KEGG, Right) and of the sections of pathways (modules, Left), as measured by the absolute value of normalized weights (color-coded yellow strongest to blue weakest) (see interactive version of this map in http://pathways.embl.de/metagenomics). Nodes symbolize compounds, and lines connecting nodes are enzymes. All enzymes (lines) corresponding to a single KEGG map or a single module will have the same color. Shaded gray Insets (A–G) for pathways and corresponding insets for modules (no modules available for C and D) indicate examples from the text: energy conversion (A–E), amino acid metabolism (G), and lipid synthesis and glycan metabolism (F). Photosystem I and II modules (A, bright yellow to green) show significant covariation with the environment, but the ATPase is invariant (blue). (B) A similar pattern was observed for oxidative phosphorylation (see text for more details). (C) Pieces of the photosynthetic machinery (including heme/porphyrin synthesis). (D) Carbon fixation. (F) Glycerophospholipid pathways show that only the “pipe” leading to or from the citrate acid cycle covaries. (G) Amino acid metabolic pathways discussed in the text. For map generation, see ref. 26.
Adaptation of Energy Conversion Strategies to Specific Environmental Challenges.
Many of the environmentally-dependent pathways were associated with energy conversion. The diversification in energy-conversion strategies is reasonable given that a primary challenge to all microbial communities is how to maintain adequate energy reserves despite challenging conditions in their specific environment.
Our results demonstrate ample diversification in energy-conversion strategies linked to such quantitative environmental differences. In particular, we show that proteins involved in (photo)autotrophic processes, such as photosynthesis, oxidative phosphorylation, and carbon and nitrogen fixation, are strongly influenced by variation in environmental parameters (Fig. 3). This link is seen at all functional levels and reinforced by multiple methodologies (Table S7). The module-level analysis showed that only photosynthetic modules involved in light capture and electron transport (photosystem I, II, and the cytochrome b6/f complexes) correlated with the environment. In contrast, the abundance of the module for the ATP synthase complex, whose function is independent of the particular energy conversion strategy, does not change significantly (Fig. 3A). A similar trend can be seen for oxidative phosphorylation, although not as strongly (Fig. 3B). The seeming lack of environmental constraint on the ATP synthases probably reflects their role in coupling energy to a proton gradient (e.g., oxidative phosphorylation, etc.) that are required regardless of which specific energy-conversion strategy is used. Also, in some cases, our approach allows the 3-way linking of functional, phylogenetic, and environmental patterns. For example, in respiratory complex I, the module covering the cyanobacterial NADH dehydrogenases (i.e., most likely those from Prochlorococcus-like species) covaries positively with temperature and other photosynthesis modules (Tables S3 and S4). However, the module covering the proteobacterial NADH dehydrogenase (i.e., most likely from SAR11-like species) varies inversely with the temperature gradient. Such observations can be associated with their respective geographic distributions. Photosynthetic Prochlorococci are mostly absent in the northern, temperate sites but dominate in tropical waters (10, 12); whereas SAR11-like proteobacteria, which do not rely on the classical photosynthetic machinery to collect energy, dominate the northern, temperate regions (13). Thus, although variation in the reliance on autotrophic processes is not unexpected, these observations illustrate the potential of the proposed methodology to detect biologically relevant covariation.
Balancing Amino Acid Synthesis vs. Import: Adapting to Nutrient-Limited Conditions.
We observed that metabolic pathways associated with amino acid and cofactor transport and metabolism varied significantly with the environmental features. Given the oligotrophic nature of the oceans (14), this observation may reflect the variability in amino acid uptake and recycling pathways as an alternative nutrient source in the various environments sampled, a strategy used by many of the dominating species in ocean surface waters (15). Lending further support to this hypothesis, operons with significant structural correlations consisted of both amino acid metabolism pathways and transporters necessary for exogenous uptake (Table S8). Amino acid uptake is sensitive to light availability (15), which, given the north to south sample collection gradient, could be an additional factor in their variation. The strength of this covariation is further reflected by the positioning of many of the amino acid metabolism maps along the same principal axis as temperature and chlorophyll in the positive direction (Fig. S2).
Environmentally Variant/Invariant Amino Acid Pathways Differ by Cofactor Cost.
One of the most striking aspects of our findings is that amino acid biosynthetic pathways could be divided into those that vary with the environment (high structural correlation coefficient) and those that do not. Interestingly, covariation of amino acid biosynthesis with the environment was unrelated to the energetic cost of synthesizing a particular amino acid (e.g., metabolic optimization). This simple result is seemingly counterintuitive, as one would expect that those pathways that used the most energy might vary the most with the energetic potential of their environment. However, we observe a significant positive correlation (P < 0.05) between the structural correlation of the amino acid pathways (strength of environmental covariation) and their dependence on potentially limiting cofactors (e.g., thiamin, tetrahydrofolate, cobalamin; see Materials and Methods and Table S9), corroborated by concordant variation in the ABC transporters of the cofactors.
This result suggests that the “cost” of obtaining trace metals for use in cofactors could be more expensive than the energetic cost of synthesizing transport machinery and degradative components that would allow for import of exogenous amino acids reducing the need for cofactor. The relationship among an environmental covariation of an amino acid, cofactor dependency, and transporters suggests the idea of “synthesis vs. import” as an adaptive strategy in aquatic environments; i.e., the import of exogenous amino acids may be more favorable than direct synthesis in environments where the manufacture of the cofactors required for their synthesis is limiting.
Environment-Driven Variation in Methionine-Dependent Pathways.
Methionine, a central amino acid in oceanic microorganisms, presents a particularly interesting example of this phenomenon and, also, illustrates the importance of a complex network of metabolic adaptations to limiting factors. Reduction in the use of methionine in nutrient limited environments has been noted previously (16). Our results suggest this reduction may stem from cofactor (and perhaps more specifically metal) cost optimization rather than (or in addition to) energetic constraints. We find environmentally-linked variation throughout methionine metabolism, including methionine synthesis, salvage, and degradation reinforced at multiple levels of pathway resolution. More specifically, we note that synthesis of both methionine and its cofactor cobalamin (contains cobalt) both decrease as methionine degradation and amino acid transporters (e.g., spermidine and putrescine) increase. Oceanic microorganisms have been shown to take extreme measures to conserve limited metals (e.g., iron) (17); these observations suggest an analogous adaptive response to cobalt limitation.
If such a limitation exists, one would expect to find equally wide-spread changes throughout methionine- (and, thus, cobalamin-) dependent pathways; in particular, in those that depend on the cofactor S-adenosylmethionine (SAM), such as methylation and secondary metabolites biosynthesis. Indeed, we do find evidence for environmental dependence for a whole suite of methionine processes, including cobalamin biosynthesis, as well as variation in many of the SAM-dependent processes (e.g., polyamines, ubiquinone, chlorophyll, and heme), hinting that methionine has a significant role in shaping downstream environmental adaptations. These observations provide evidence in support of a synthesis vs. import theory.
Modulating Lipid and Glycan Metabolism As an Adaptation to Physicochemical Conditions.
Lipids and glycans are important components of the microbial outer membrane; thus, it would be expected to be particularly responsive to environmental conditions. We do find strong environment-linked variation in a plethora of lipid and glycan metabolism-related processes (see Fig. 3 and Table S10). Indeed, modification of the cell wall is a known adaptive mechanism (e.g., for membrane fluidity) (9, 18), and the variation of pathways involved in extracellular polysaccharide synthesis, lipopolysaccharide synthesis, cell wall maintenance, and glycerophospholipid synthesis (Table S4) along the salinity, sample depth, and temperature gradients sampled in the GOS sites could be a reflection of this adaptation. Also, significant contributions of lipid metabolism modules in the construction of a LM for sample depth (Table S4) may illustrate an adaptation strategy to maintain buoyancy for optimal growth conditions (e.g., to optimally profit from light scavenging machinery adaptations for certain wavelengths, see ref. 12). Alternatively, it could reflect an adaptation of heterotrophic prokaryotes to the varying composition of phytoplankton-produced dissolved organic matter with depth. Due to the diversity of these roles of pathways without further experimentation, one can only speculate on the validity of these particular interpretations. However, undoubtedly, the extreme variation and flexibility of these pathways indicate their central importance in metabolic adaptation to the environment.
Discussion
As different evolutionary strategies are required to cope with the unique set of challenges specific to each geographic site sampled, our results suggest how environmental pressures shaped these pathway differences. The detailed analysis of 3 case studies revealed particular pathway adaptations that provide numerous testable hypotheses for linking metabolic versatility to the environment.
Recently, Dinsdale et al. (7) demonstrated that functional differences can be used to discriminate among 9 qualitatively categorized, discrete ecosystems. However, as in genome wide association studies where methods using binned data have been supplemented by more sensitive methods that make use of continuous measurements (19), we have demonstrated the utility of a similar transition in microbial ecology by using comparative metagenomics. Our methods associate microbial community functions with quantitative, continuous features of the environment, allowing for an objective, data-driven framework to classify sites both on the basis of their metabolism and environmental parameters. We show evidence for widespread environmentally-dependent metabolic versatility even in seemingly similar sites (sharing same habitat classification). The methods implemented here also provide a valuable and sensitive assay for simultaneously assessing a number of environmental parameters, allowing us to predict both individual and groups of environmental features (see Fig. 2 and Table S4). In reverse, we also predict the usage of a particular metabolic pathway given a set of environmental conditions (Table S11). Thus, our results suggest that metabolic footprints can be used as the basis of biosensors in situations where no clear measurable environmental factors are available (e.g., monitoring water quality and predicting health state from clinical samples). Indeed, such biosensors would provide more information than the current practice based on species composition (20), which measure downstream effects (e.g., marker species in pollution), instead of focusing on the molecular processes of the ecosystem as a whole.
Like all current metagenomics datasets, the GOS dataset provides only a snapshot of the total genomic content of a site. However, by quantifying the difference in pathway usage along different environmental gradients, one can observe the SPATIAL dynamics of pathways—analogous to the temporal dynamics in usage of pathways between different cellular states (27).
Although we have taken precautions to ensure the coverage across sites is the same (SI Materials and Methods), the potential remains for important but rare components of metabolic adaptation to be overlooked. Similarly, although we were able to map 74% of proteins to STRING OGs, there is still a fraction of hypothetical proteins that may harbor unknown and, thus, “unmapped” metabolic components. Indeed, environmental covariation may provide contextual clues for the annotation of proteins of unknown function. Novel techniques to functionally characterize this fraction represent a significant challenge and an avenue of active research (21, 22). Also, the 5 features reported do not fully encapsulate environmental complexity, and the integration of more environmental measurements will likely reveal many new and exciting discoveries. Despite the inherent limitations of the data, they do not compromise the ideas or the conceptual framework presented. Indeed, although the available datasets have constrained us to an analysis of aquatic habitats, the same methodology could readily be applied to investigate the specific metabolic capabilities for any ecosystem in which (physical) environmental parameters are collected including (e.g., different anatomical locations, which form “microbial habitats” in humans).
The potential contribution of large viruses as a reservoir for microbial diversity has recently been shown (23, 24). However, <0.3% of proteins in our set can be characterized as viral, suggesting a negligible impact on our reported findings (see Materials and Methods). Repeating this analysis on just the viral sample represents an interesting avenue for future research.
It is clear that microbial communities have a critical role in shaping our world from aiding in global climate regulation (25) and geochemical cycles to degrading hazardous byproducts; however, the complicated, intertwined nature between microbial communities and the environments they inhabit and influence remains poorly understood. We have presented a methodological framework that provides a roadmap to explore these questions in a systematic and statistically rigorous fashion.
Materials and Methods
General Overview of Methodology and Data Used.
For full data and code dump, see http://networks.gersteinlab.org/metagenomics. For full details and extensive discussion of the validation of chosen procedures, see SI Materials and Methods.
Preprocessing GOS Data.
Sequence and metadata (Fig. S3) from 37 sites (Table S1) from the first phase of the GOS expedition (0.1- to 0.8-μm filter size (i.e., mostly prokaryotes), except for Sargasso Sea station 11; see ref. 35) was downloaded from CAMERA (36).
Mapping.
Peptides were mapped to sites (Figs. S4 and S5) based on the read-to-scaffold and orf-to-scaffold mappings available at CAMERA (36). Then, 111 KEGG maps, 141 modules, and 191 operons were assigned as indicated in ref. 14. Module definitions were downloaded from KEGG (37), and operons were constructed as indicated in ref. 38. In brief, protein sequences were searched against the extended database of proteins assigned to OGs in STRING 7.0 (38) by using BLASTP (39). A pathway was called present when a hit matched any of its components (bitscore >60; 80% consistency among top 5 hits; see SI Materials and Methods). Cofactors were mapped to each module by means of EC number by using the Brenda database (40). All results described were manually scrutinized to reduce artefactual assignments. The pathway frequency for each site was calculated by summing the total number of instances of that pathway in a particular site, then normalized by total number of assignments for all pathways in that site to compensate for sample coverage differences. Further normalizations were performed when necessary (see SI Materials and Methods). For all analyses, pathways for which the summed count over all sites constituted equal to or <0.01% of the total count were removed to avoid artifacts.
Correlation and Regression.
We computed pairwise Spearman correlations between each pathway frequency vector and each environmental metadata vector. Linear regression models were constructed in 2 directions: (i) the environmental factor was treated as the response variable and predicted from a subset of pathway frequencies; and (ii) the inverse model where pathway frequency was treated as the response variable and predicted from environmental factors (see SI Materials and Methods).
DPM.
DPM was used to analyze whether groupings of sites based on similar environmental features also shared functional similarities. Sites were clustered based on their quantitative environmental metadata into “site-sets.” Next, we partitioned the sites in the metabolism matrices (Fig. 1A) into the same site sets and tested whether the means of each individual map, module, operon, and COG between the 2 site sets differed significantly (Benjamini–Hochberg corrected P < 0.05). Significant pathways were combined to form the DPM footprint (Table S5).
Acknowledgments.
We thank Andrea Sboner, Roger Alexander, Albert Colman, and Nick Ornston for many thoughtful discussions, and Minoru Kanehisa's team for early access to the KEGG module database. The instrumentation was supported by the Yale University Biomedical High Performance Computing Center and National Institutes of Health Grant RR19895. J.O.K. was supported by a Marie Curie fellowship and J.R. and P.B., by the European Union FP7 Program Contract no. HEALTH-F4–2007-201052.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808022106/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0808022106
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2629784?pdf=render
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/full/106/5/1374
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/content/abstract/106/5/1374
Free after 6 months at www.pnas.org
http://www.pnas.org/cgi/reprint/106/5/1374.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0808022106
Article citations
EMPathways2: Estimation of Enzyme Expression and Metabolic Pathway Activity Using RNA-Seq Reads.
Methods Mol Biol, 2812:39-46, 01 Jan 2024
Cited by: 0 articles | PMID: 39068356
Chronic industrial perturbation and seasonal change induces shift in the bacterial community from gammaproteobacteria to betaproteobacteria having catabolic potential for aromatic compounds at Amlakhadi canal.
World J Microbiol Biotechnol, 40(2):52, 26 Dec 2023
Cited by: 0 articles | PMID: 38146029
DNA sequencing, microbial indicators, and the discovery of buried kimberlites.
Commun Earth Environ, 4(1):387, 21 Oct 2023
Cited by: 0 articles | PMID: 38665197 | PMCID: PMC11041713
Diversity and salinity adaptations of ammonia oxidizing archaea in three estuaries of China.
Appl Microbiol Biotechnol, 107(22):6897-6909, 13 Sep 2023
Cited by: 1 article | PMID: 37702790
Functional comparison of metabolic networks across species.
Nat Commun, 14(1):1699, 27 Mar 2023
Cited by: 3 articles | PMID: 36973280 | PMCID: PMC10043025
Go to all (107) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held October 11-13, 2002, in Longboat Key, Florida
American Society for Microbiology, Washington (DC), 30 Oct 2020
Cited by: 0 articles | PMID: 33119236
ReviewBooks & documents Free full text in Europe PMC
Merging metagenomics and geochemistry reveals environmental controls on biological diversity and evolution.
BMC Ecol, 14:16, 28 May 2014
Cited by: 11 articles | PMID: 24886397 | PMCID: PMC4047435
Functional Responses of Salt Marsh Microbial Communities to Long-Term Nutrient Enrichment.
Appl Environ Microbiol, 82(9):2862-2871, 18 Apr 2016
Cited by: 13 articles | PMID: 26944843 | PMCID: PMC4836423
Parallel Evolution of Genome Streamlining and Cellular Bioenergetics across the Marine Radiation of a Bacterial Phylum.
mBio, 9(5):e01089-18, 18 Sep 2018
Cited by: 16 articles | PMID: 30228235 | PMCID: PMC6143742
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
Development of a graph-theoretic approach to predict protein function by integrating large scale heterogeneous data
Dr Paccanaro, Royal Holloway, Univ of London
Grant ID: BB/F00964X/1
European Commission FP7 (1)
Grant ID: FP7_201052
NCRR NIH HHS (2)
Grant ID: RR19895
Grant ID: S10 RR019895





