Abstract
Free full text

SERIES: Genomic instability in cancer Balancing repair and tolerance of DNA damage caused by alkylating agents
Abstract
Alkylating agents comprise a major class of frontline chemotherapeutic drugs that inflict cytotoxic DNA damage as their main mode of action, in addition to collateral mutagenic damage. Numerous cellular pathways, including direct DNA damage reversal, base excision repair (BER), and mismatch repair (MMR) respond to alkylation damage to defend against alkylation-induced cell death or mutation. However, maintaining a proper balance of activity both within and between these pathways is crucial for an organism's favorable response to alkylating agents. Furthermore, an individual's response to alkylating agents can vary considerably from tissue to tissue and from person to person, pointing to genetic and epigenetic mechanisms that modulate alkylating agent toxicity.
Introduction
Alkylating agents are a ubiquitous family of reactive chemicals that transfer alkyl carbon groups onto a broad range of biological molecules, thereby altering their structure and potentially disrupting their function. Alkylating agents are practically unavoidable due to their abundant presence in the environment and within living cells. Major sources of external alkylating agents include constituents of air, water and food such as biological byproducts (e.g. abiotic plant material) and pollutants (e.g. tobacco smoke and fuel combustion products)1-3. Internally, alkylating agents can arise as byproducts of oxidative damage or from cellular methyl donors such as S-adenosylmethionine, a common cofactor in biochemical reactions4, 5. Due to the cytotoxic, teratogenic and carcinogenic effects caused by alkylation damage, alkylating agents pose significant threats to human health6. In spite of this, certain toxic alkylating agents are commonly used systemically, as chemotherapeutic drugs in cancer patients, with the goal of killing cancer cells7. Consequently, while alkylating agents can induce cancer, they are also used to treat cancer. Based upon the double-edged properties of alkylating agents, a greater understanding of the cellular factors that determine biological outcome in response to alkylation damage is particularly relevant for both cancer prevention and cancer therapy, in addition to general human health. The biological response to alkylating agents can be quite complex due to the variety of lesions introduced by a single alkylating agent in combination with the diversity of cellular repair mechanisms and response pathways that can be elicited upon alkylation damage. Here, we focus on the interplay between the multiple cellular factors that respond to DNA alkylation damage and how they collectively determine sensitivity or resistance to alkylating agents.
Molecular damage caused by alkylating agents
Alkylating agents react with the ring nitrogens (N) and extracyclic oxygen (O) atoms of DNA bases to generate a variety of covalent adducts ranging from simple methyl groups to complex alkyl additions8, 9 (FIG. 1 and and2).2). The pattern of DNA lesions generated by an alkylating agent depends on the number of reactive sites within the alkylating agent (monofunctional versus bifunctional), its particular chemical reactivity (SN1 versus SN2-type nucleophilic substitution), the type of alkyl group addition (methyl, chloroethyl, etc.) and the DNA substrate (double- or single-stranded). Monofunctional alkylating agents contain one active chemical moiety for modification of a single site in DNA whereas bifunctional alkylating agents contain two reactive groups that can bond with separate DNA bases to form interstrand crosslinks. Whereas SN2-alkylating agents mainly target ring nitrogen atoms in DNA bases, SN1-alkylating agents can modify these nitrogens plus the extracyclic oxygen groups (FIG. 1). Notably, nearly all chemotherapeutic alkylating drugs currently used in the clinic are SN1-type alkylating agents and they can be either monofunctional or bifunctional (FIG. 2).
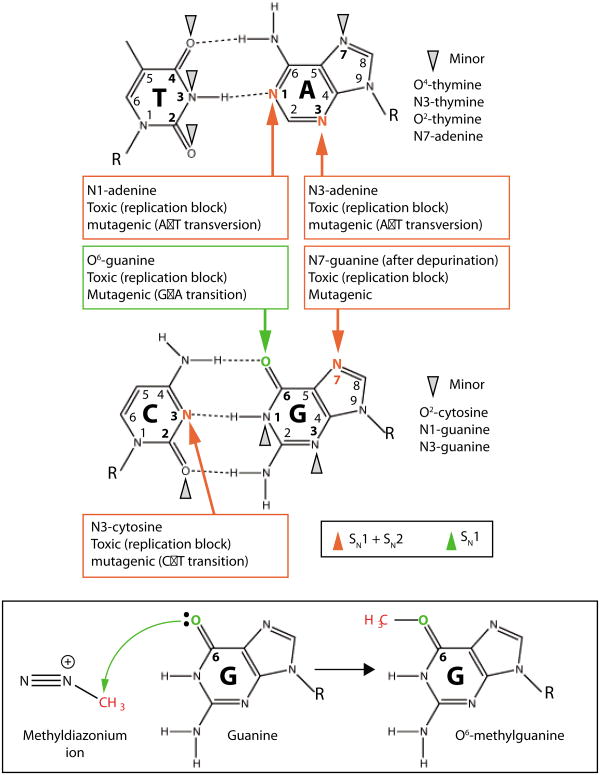
a) Alkylating agents react with the nitrogen and oxygen atoms of DNA bases to form covalent alkyl lesions. The major sites of alkylation on the DNA bases and biological effects of alkylation are noted by red and green arrows with minor lesions denoted by grey arrow heads. b) An example of a DNA alkylation reaction between the methyldiazonium ion of the chemotherapeutic alkylating agent, temozolomide, with the O6-position of guanine to form the O6-methylguanine DNA lesion. R, ribose-phosphate DNA backbone.
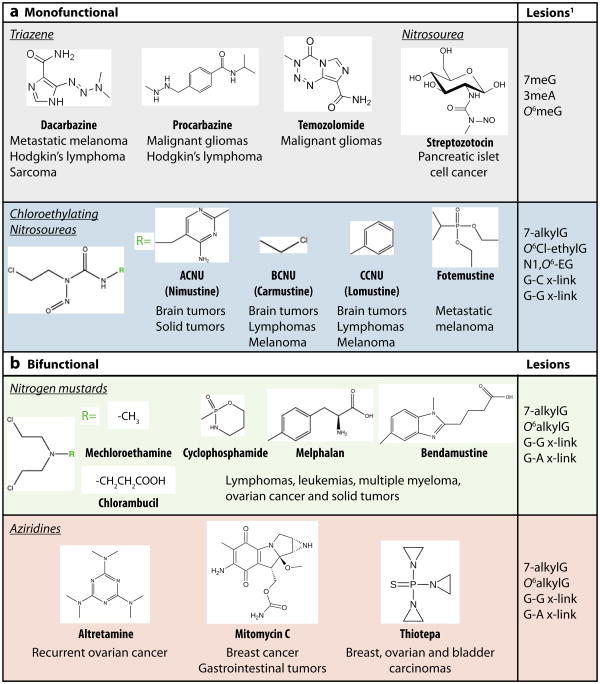
Monofunctional methylating agents (part a) such as temozolomide induce the formation of 7-methylguanine (7meG), 3-methyladenine (3meA) and O6-methylguanine (O6meG) as the major DNA lesions. The O6meG adduct can readily mispair with thymine to form O6meG:T mispairs. In single-stranded DNA, monofunctional alkylating agents can generate 1-methyladenine (1meA) and 3-methylcytosine (3meC) as well. Monofunctional chloroethylating agents induce the formation of O6-chloroethyl guanine (O6Cl-ethylG) as the biologically relevant lesion. The O6Cl-ethylG is prone to cyclization to form N1,O6-ethanoguanine (N1,O6-EG) which can react with another guanine to form a guanine-guanine (G-G) interstrand crosslink (x-link). Bifunctional alkylating agents (part b) such as BCNU have two reactive sites that can crosslink two strands of DNA to form interstrand DNA crosslinks as well.
Due to the high nucleophilic reactivity of the N7-position of guanine in DNA, most monofunctional methylating agents induce the formation of N7-methyl guanine (7meG) as the predominant methylation adduct, accounting for 60-80% of the total alkylation lesions in DNA (FIG. 1). By itself, 7meG does not possess any mutagenic or cytotoxic properties but it is prone to spontaneous depurination to form apurinic/apyrimidinic (AP) sites that are toxic and mutagenic. In addition to 7meG, monofunctional methylating agents can generate N3-methyladenine (3meA) as the other primary N-methylation product, accounting for 10-20% of total methyl adducts9. In contrast to the relatively innocuous 7meG lesion, the 3meA lesion is highly cytotoxic since it can block most DNA polymerases and thereby inhibit DNA synthesis10, 11. In single-stranded DNA, the N1-position of adenine and the N3-position of cytosine are also subject to methylation by monofunctional methylating agents to generate the replication blocking and mispairing lesions, 1-methyladenine (1meA) and 3-methylcytosine (3meC)8 (FIG. 1). In double-stranded DNA, these sites are protected due to base pairing, but they can be transiently exposed during replication, transcription or recombination.
Amongst the oxygen atoms of DNA, the O6-position of guanine represents a major site of methylation by SN1-type alkylating agents to generate O6-methylguanine (O6meG) (FIG. 1). Even though O-alkyl lesions are generated to a much lesser extent than N-alkyl adducts, the induction of O6meG lesions by alkylating agents is of great biological relevance because O6meG can readily mispair with thymine during DNA replication to cause many of the mutagenic and cytotoxic biological effects of alkylating agents (discussed in detail below). Alkylating agents can also modify other nitrogen and oxygen atoms in DNA besides the aforementioned sites to generate additional toxic and mutagenic lesions (FIG. 1,). However, these lesions are observed at 10 to 100-fold lower levels and represent a minor fraction of total alkylation adducts.
A number of monofunctional SN1-methylating agents are currently used as anticancer drugs; these include the triazine family of compounds such as dacarbazine, procarbazine and temozolomide as well as the nitrosourea compound, streptozotocin7 (FIG. 2). Due to their chemical reactivity as monofunctional SN1methylating agents, these chemotherapeutic alkylating agents produce significant levels of 7meG, 3meA and O6meG lesions as the primary alkylated DNA adducts. The chloroethylating agents represent another major class of monofunctional alkylating agents that react with DNA with a similar specificity as SN1-methylating agents except with the addition of a chloroethyl group. The majority of nitrosourea compounds used in the clinic are chloroethylating agents that can modify the N7 and O6-positions of guanine to generate chloroethyl adducts (FIG. 2). Significantly, O6-chloroethyl guanine (O6Cl-ethylG) adducts undergo rapid chemical rearrangement to react with nearby cytosine bases to generate guanine-cytosine (G-C) interstrand DNA crosslinks that are highly cytotoxic.
Bifunctional alkylating agents have similar reactive properties to monofunctional alkylating agents but contain two active moieties that can react with separate bases of DNA to form interstrand crosslinks in addition to monoadducts (FIG. 2). The nitrogen mustards and aziridine compounds are two major classes of bifunctional alkylating drugs used for cancer treatment that can crosslink DNA through a aziridinium-ring intermediate12. Nitrogen mustard compounds react readily with N7-guanine and to a lesser extent, N3- and N7-adenine, to form bulky N-monoadducts. These adducts can subsequently react with another base to form guanine-guanine (G-G) and guanine-adenine (G-A) interstrand crosslinks (FIG. 2). The nitrogen mustard, mechloroethamine, was the first chemotherapeutic drug used on cancer patients, and derivatives thereof (chlorambucil, cyclophosphamide, melphalan and bendamustine) are prescribed for the treatment of a wide variety of cancers. Aziridine compounds such as altretamine, mitomycin C, and thiotepa use a reaction similar to nitrogen mustards to form G-G or G-A interstrand crosslinks in addition to a variety of monoadducts.
Depending on their position in DNA, the different base adducts introduced by alkylating agents can compromise genome integrity by inducing mutagenesis (thereby promoting cancer induction) and/or blocking essential biological processes such as DNA replication and transcription (potentially leading to cell death) (FIG. 1). Moreover, certain lesions can also be processed into clastogenic and cytotoxic products that can engage other DNA repair pathways or induce programmed cell death. Although we will focus on the biological effects of alkylation damage to genomic DNA, it is important to note that other biological molecules are subject to alkylation damage, including RNA, protein, lipids, and mitochondrial DNA. Thus, a single chemotherapeutic alkylating agent is able to modify a variety of biological molecules to generate a spectrum of lesions that can elicit a number of biological effects.
Complex cellular responses to DNA alkylation damage
Just as a single alkylating agent can cause multiple types of alkylated base lesions, each lesion can be repaired or processed by a number of enzymes and pathways in an effort to counteract the genomic damage induced by alkylating agents, and to protect an organism from alkylation toxicity and mutagenesis (FIG. 3). Thus, diverse cellular repair pathways collectively modulate alkylation sensitivity. This occurs through an overlap in substrates, compensating pathways, processing of intermediates by alternate pathways, or direct crosstalk between pathways. The major repair mechanisms for alkylation damage include direct DNA repair by the AlkB dioxygenase enzyme and the O6-methylguanine-DNA methyltransferase (MGMT) repair protein; and by the multistep pathways of base excision repair (BER) and nucleotide excision repair (NER) (FIG. 4). The AlkB enzyme catalyzes direct reversal of certain N-alkyl lesions (e.g. 1meA and 3meC) through a unique oxidative dealkylation reaction, whereas MGMT directly repairs O6meG and O6Cl-ethylG lesions via transfer of the alkyl group to its active site cysteine residue13. The BER pathway plays an important role in the repair of many N-alkyl lesions (e.g. 3meA and 7meG), and the NER pathway may also contribute to the repair of alkylated base lesions14, 15, although this pathway is primarily thought to function in the repair of bulkier base lesions that significantly disrupt DNA-helix structure (e.g. intrastrand crosslinks and UV-induced DNA damage)16.
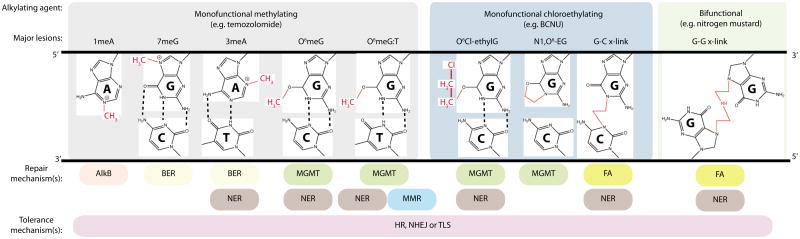
The major repair and tolerance mechanisms for each lesion are listed below each adduct. See text and Figure 3 for more details on the repair mechanisms. Green lines indicate repair-dependent DNA synthesis; red structures are alkyl modifications. BER, base excision repair; FA, Fanconi anaemia; MMR, mismatch excision repair; MGMT, O6-methylguanine DNA methyltransferase; NER, nucleotide excision repair.
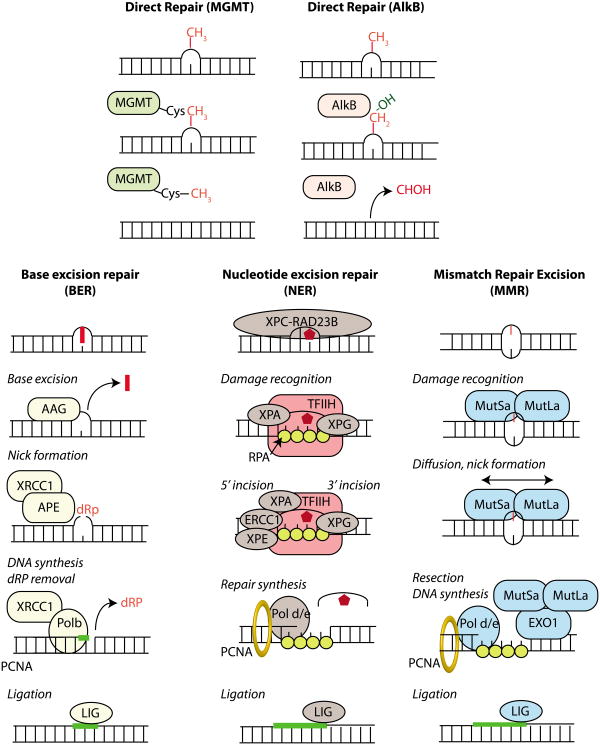
a) Direct repair results in the reversal of an alkylated base to a normal base without any intervening steps or the generation of DNA strand breaks. The O6-methylguanine DNA methyltransferase (MGMT) catalyzes the transfer of the aberrant methyl group on the DNA base onto an active site cysteine residue in MGMT, thereby removing the methyl lesion from DNA. The AlkB family of dioxygenases catalyze direct DNA damage reversal through hydroxylation (-OH addition) of the methyl group on the damaged DNA base; the hydroxylated methyl group then spontaneously leaves as formaldehyde (HCOH) to regenerate the normal base. b) The base excision repair (BER) pathway is responsible for removing simple alkyl and oxidative base lesions. BER is initiated by a DNA glycosylase that excises the damaged base from DNA to generate an abasic site for subsequent processing by either short-patch or long-patch BER. In short patch BER (shown here), an apurinic/apyrimidinic endonuclease (APE) incises the abasic site to yield a 3′-hydroxyl adjacent to a 5′-deoxyribosephosphate (5′dRP) moiety. DNA Polymerase β (Pol β) removes the 5′dRP residue via its lyase domain and extends from the 3′OH with its polymerase domain generating a nick ready for subsequent ligation by DNA ligase (LIG). Long patch repair occurs in a similar manner except that a longer piece of DNA is removed. c) Nucleotide excision repair (NER) is responsible for removing bulky, helix-distorting lesions but can serve as a backup repair mechanism for the excision of certain alkylated bases16. NER is initiated by the recognition of a distorted region containing the DNA adducts followed by 5′ and 3′ incision on either side of the lesion to create a 27-29 nucleotide gap. The excised region is then filled in by DNA Pol δ or ε for subsequent ligation by DNA ligase. d) The mismatch excision repair (MMR) pathway mediates the removal of mismatched bases as well as mispairs caused by base alkylation lesions such as O6meG:T. In mammals, MMR is initiated by the recognition of mismatches by the MutSα heterodimer (MSH2-MSH6) followed by recruitment of the MUTLα heterodimer (PMS2-MLH1). Diffusion of MUTSα-MUTLα leads to nicking of the DNA either upstream or downstream of the mismatch that serves as an entry point for exonuclease 1 (EXO1) that removes a segment of DNA, which is subsequently filled in and repaired by a combination of Pol δ, ε and DNA ligase to complete repair.
Numerous additional repair pathways play significant roles in the cellular response to alkylating agents by processing or bypassing the downstream DNA lesions resulting from DNA transactions at the primary alkyl lesion (e.g. DNA mispairs and replication blocks) or repairing secondary lesions (e.g. single and double strand DNA breaks and crosslinks) (FIG. 3 and and4).4). The mismatch excision repair (MMR) pathway recognizes and processes DNA base mismatches to remove misincorporated nucleotides; this includes the O6meG:T mispairs that arise by the incorporation of thymine opposite O6meG during replication of alkylated DNA17. Both the homologous recombination (HR) and non-homologous end joining (NHEJ) pathways repair DNA double strand breaks (DSBs); HR uses homologous DNA (sister chromatids) as a template to resynthesize DNA over the break resulting in sister chromatid exchanges (SCEs), whereas NHEJ joins the DNA ends with no or little homology. The translesion DNA synthesis (TLS) pathway employs low-fidelity DNA polymerases to bypass lesions that stall high-fidelity replicative polymerases, thereby alleviating any blocks during DNA replication. Importantly, HR generally results in error-free repair whereas NHEJ and TLS are usually error-prone repair mechanisms that can give rise to mutations. Finally, the Fanconi anaemia pathway coordinates elements of NER, HR and TLS to function in the replication-dependent repair of interstrand DNA crosslinks caused by particular alkylating agents. Due to their widespread involvement in the response to DNA alkylation damage, we will highlight the roles of these DNA repair mechanisms throughout this manuscript while referring readers to more comprehensive reviews dedicated to these pathways18-20.
With the abundance of repair pathways that act upon DNA alkylation damage, it has become clear that an imbalance in any one pathway can affect the overall cellular response, resulting in dramatic effects on the alkylation sensitivity of a cell, a tissue, or a whole organism. Moreover, pathways such as BER and NER require the coordination of multiple enzymatic steps in order to be completed accurately. If an imbalance occurs such that the steps lose coordination, toxic intermediates accumulate that often exhibit greater toxicity than the original DNA base lesion. Furthermore, just as imbalances can occur within a particular DNA repair pathway, they can also occur between different pathways. In this Review, we focus on the distinct and overlapping pathways involved in the repair of 3meA and O6meG, two representative DNA lesions induced by many cancer chemotherapeutic agents.
Deleterious consequences of BER imbalance
BER is a highly coordinated, multi-step pathway that removes a damaged DNA base (such as an alkylated base) and ultimately replaces it with the correct base (FIG. 3 and and5).5). The initiation of BER occurs by the recognition and excision of a damaged DNA base lesion by a DNA glycosylase. In humans, there are currently 11 known DNA glycosylases21. Here we will focus on the alkyladenine-DNA glycosylase (AAG, also known as MPG or ANPG) as it is responsible for excising the cytotoxic 3meA DNA lesion induced by both SN1 and SN2 alkylating agents.
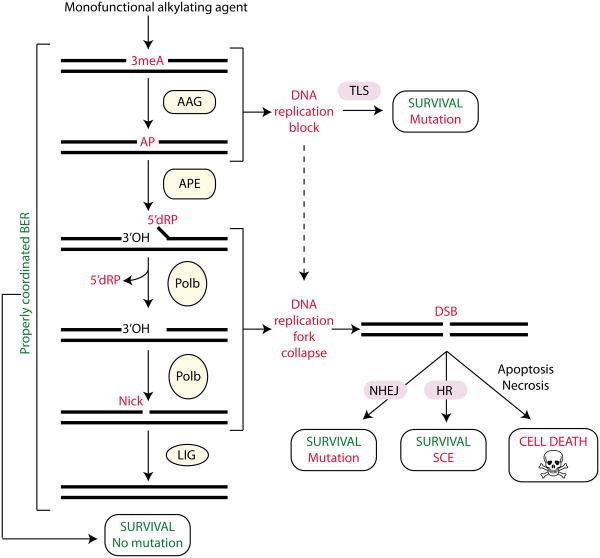
Monofunctional methylating agents induce the formation of 3-methyladenine (3meA) lesions, which are excised by the alkyladenine DNA glycosylase (AAG) to generate an apurinic/apyridinic site (AP). The apurinic/apyridinic endonuclease (APE) incises the DNA backbone immediately 5′ to the AP site generating a single strand break (SSB) containing a 3-hydroxyl (3′OH) and a 5′-deoxyribose phosphate (5′dRP). DNA polymerase β (Pol β) removes the 5′dRP moiety through intrinsic lyase activity and fills in the resulting gap. The nick is then sealed by DNA ligase (LIG) to complete base excision repair (BER). If BER is inhibited or downstream steps of BER are limiting, then toxic intermediates can accumulate leading to DNA replication blocks, replication fork collapse, double strand breaks (DSBs) and eventually cell death. The translesion DNA synthesis (TLS), homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways can bypass or repair secondary damage caused by alkyl lesions to promote cell survival at the expense of mutation or sister chromatid exchange (SCE).
Importantly, every step of BER generates intermediates [AP sites, 5′-deoxyribose phosphate (5′dRP) residues and single strand breaks (SSBs)], which have been shown to be both mutagenic and toxic (FIG. 3 and and5).5). In fact, the BER intermediates are often more toxic than the initiating DNA base lesion (in this case 3meA, a replication-blocking lesion) presumably because translesion polymerases are capable of bypassing the unrepaired 3meA lesion with varying efficiency and accuracy10, 22-24. Thus, it is essential for BER to be tightly controlled to avoid an accumulation of any of these toxic intermediates. Indeed, another key BER protein, XRCC1, coordinates the DNA processing events of BER by interacting with each of the aforementioned DNA processing enzymes to ensure the proper completion of BER. Accordingly, it has been proposed that the BER pathway functions similar to “passing the baton” in a relay race; one enzyme complex passes the repair intermediates on to the next enzyme complex, essentially sequestering the intermediates and preventing their toxicity25, 26. Due to the tight coupling of BER processing, an imbalance in any step can therefore alter the phenotypic response to alkylating agents; imbalances can occur when a BER enzyme exhibits an alteration in activity or expression level without compensating changes in the downstream steps of the BER pathway. Although single nucleotide polymorphisms (SNPs) found in BER genes have been associated with an increased risk of multiple types of cancer, additional studies are required to determine the functional consequences and the significance of these SNPs in cancer patients (TABLE 1). However, the potential for imbalanced BER in human cancer is exemplified by the colorectal cancer predisposition syndrome, MAP (MYH-associated polyposis), a consequence of biallelic-inherited mutations in the DNA glycosyslase, MYH27, 28 (TABLE 1).
Table 1
| DNA repair protein | Alteration | Cancer | References |
|---|---|---|---|
| BER | |||
 AAG AAG | Single nucleotide polymorphisms and altered expression | Osteosarcoma, breast cancer and astrocytic tumors | 44-47 |
 MYH MYH | Autosomal recessive mutations | Colorectal cancer | 27,28 |
 APE1 APE1 | Altered expression or localization | Numerous cancers | 78-83 |
 Pol β Pol β | Single nucleotide polymorphisms | Numerous cancers | Reviewed in 84 |
 XRCC1 XRCC1 | Single nucleotide polymorphisms | Numerous cancers | Reviewed in 85 |
| MGMT | Increased expression and activity | Multiple cancers, glioblastoma, pediatric brain tumors | Reviewed in 127 |
| MMR | Loss of function mutation, loss of expression | Colorectal, endometrial, gastric and urothelial cancers | Reviewed in 186 |
 MLH1 MLH1 | |||
 MSH2 MSH2 | |||
 MSH3 MSH3 | |||
 MSH6 MSH6 | |||
 MLH3 MLH3 | |||
 PMS2 PMS2 | |||
| HR | Loss of function mutation | Breast, ovarian, fallopian tube, prostate and pancreatic cancer, malignant melanoma, Fanconi Aneamia | Reviewed in 184, 187 |
 BRCA1 BRCA1 | |||
 BRCA2 BRCA2 | |||
 BRIP1 BRIP1 | |||
 PALB2 PALB2 | |||
| TLS | Autosomal recessive mutation Overexpression | Skin cancer Non-small cell lung cancer, glioma | Reviewed in 183 |
 POLH (Pol η) POLH (Pol η) | |||
 POLK (Pol κ) POLK (Pol κ) |
BER imbalance in vivo results in detrimental consequences
Modulation of DNA glycosylase activity can have profound effects if the downstream BER pathway is not properly coordinated29-32. Pioneer studies in bacteria and yeast have shown that increased DNA glycosylase levels are correlated with increased spontaneous mutagenesis and enhanced sensitivity to the alkylating agent, methyl methanesulfonate (MMS)33-36. This AAG-dependent increase in sensitivity to alkylation has been recapitulated in numerous mammalian cell types, including breast and ovarian cancer cells, astrocytes, and mouse embryonic fibroblasts (MEFs)31, 32, 37-40. Moreover, increased expression of AAG in mammalian cells also results in increased SCEs, chromosome aberrations, inhibition of DNA replication, and a higher number of DNA breaks in response to alkylating agents41. Even in the absence of alkylating agents, increased AAG DNA glycosylase activity can lead to detrimental effects; AAG activity is correlated with microsatellite instability and increased spontaneous frameshift mutagenesis in yeast and human cells as well as in noncancerous human tissue samples42, 43. Consequently, evidence suggests that altered AAG expression may play a role in various human cancers44-47 (TABLE 1).
The generation of both an Aag-/- knockout mouse and a transgenic mouse with increased AAG levels (Aag Tg) have provided valuable models to test the consequences of BER imbalance at the whole animal level30, 48, 49 (TABLE 2). Since AAG-initiated BER can result in the generation of toxic intermediates following MMS treatment in certain wild-type cells, increased AAG activity would promote the accumulation of toxic intermediates whereas loss of AAG could protect against the formation of these intermediates. Thus, sensitivity to alkylating agents between a wild-type, Aag-/- and an Aag Tg mouse model would be predicted to correlate with AAG activity levels (as in, sensitivity to alkylating agents: Aag Tg > wild-type > Aag-/-). Indeed, this pattern of AAG-driven toxicity has been observed repeatedly in multiple tissues (TABLE 2). For example, ex vivo bone marrow cell survival assays have shown that Aag-/- myeloid precursor cells are resistant, whereas Aag Tg cells are ultra-sensitive to alkylating agents compared with wild-type cells (50, LDS unpublished data). Moreover, Aag Tg mice exhibit vastly increased MMS-induced toxicity in numerous tissues including, spleen, thymus, retina, and cerebellum when compared with wild-type or Aag-/- mice (described in BOX 1) (30, LDS unpublished data). Finally, Aag-/- mice also exhibit remarkable protection against pancreatic β-cell death after a single high-dose of the β-cell-specific alkylating agent, streptozotocin, as well as a marked delay in the development of streptozotocin-induced glucose intolerance and diabetes51, 52. Thus, in general, Aag Tg mice exhibit much greater toxicity to alkylating agents whereas Aag-/- mice are either resistant or display no dramatic difference in sensitivity compared with wild-type mice in response to alkylating agents50.
Table 2
| Mouse model | Organ | Alkylating agent | Phenotype (relative to wildtype) | References |
|---|---|---|---|---|
| Aag-/- | Whole animal | MMS | No change in sensitivity | 49, 50 |
| Retina | MMS | Resistant to retinal degeneration | 30 | |
| Brain | MAM, MMS | Resistant to cerebellar degeneration | 119, unpublished | |
| Bone marrow (ex vivo) | MMS, TMZ | Increased resistance | 50 | |
| Spleen | MMS | Increased mutagenesis | 48 | |
| Pancreas | STZ | Resistance to β-cell toxicity and diabetes | 51, 52 | |
| Colon | AOM+DSS | Increased susceptibility to colon tumors | 55, 56 | |
| Aag-Tg | Whole animal | MMS, MNU, AOM+DSS | Increased whole animal sensitivity | unpublished |
| Brain, spleen, thymus, pancreas, bone marrow | MMS | Increased cellular toxicity | unpublished | |
| Retina | MMS | Increased retinal degeneration | 30 | |
| Mgmt-/- | Whole animal | BCNU, MNU, STZ | Increased whole animal sensitivity | 120-124 |
| Brain | MAM | Cerebellar folia atrophy, decreased granule cells | 119 | |
| Lung | MNU, nitrosamine | Increased tumors | 124, 125 | |
| Spleen | MNU, TMZ | Gross atrophy, increased apoptosis, increased mutagenesis | 122, 126 | |
| Thymus | MNU | Increased lymphoma, larger tumors | 124 | |
| Bone marrow | MNU | Hypocellular, decreased leukocytes, reduced platelet count, impaired HSC reproduction, pancytopenia | 120-123 | |
| Liver | nitrosamine | Increased tumors | 125 | |
| Colon | MNU | Increased carcinoma | 55, 181 | |
| MGMT-Tg | Brain | MAM | Normal cerebellar development | 119 |
| Lung | nitrosamine | Decreased mutation, reduced tumors | 209 | |
| Skin | MNU | Decreased papillomas, reduced tumors | 112-114 | |
| Thymus | MNU | Decreased lymphoma and mutagenesis | 107-109, 115-118 | |
| Liver | nitrosamine, spontaneous | Reduced tumor formation | 106, 110 | |
| Colon | AOM+DSS | Reduced aberrant crypt foci and K-ras mutation | 111 |
Aag, alkyladenine DNA glycosylase; AOM, azoxymethane ; BCNU, 1,3-bis(2-chloroethyl)-1-nitrosourea; DSS, dextran sodium sulfate; HSC, haematopoietic stem cell; MAM, methylazoxymethanol; MGMT, O6-methylguanine DNA methyltransferase; MMS, methylmethane sulfonate; MNU, methylnitrosourea; STZ, streptozotocin; TMZ, temozolomide.
Although complete inhibition of BER initiation (as observed in Aag-/- mice) protects against alkylation-mediated cell death in numerous tissues, it is important to note that BER inhibition does not confer protection to all alkylating agents or cell types. For example, Aag-/- embryonic stem cells as well as MEFs with reduced levels of AAG are actually sensitized to alkylating agents49, 53, 54. Furthermore, Aag-/- mice are more susceptible than wild-type mice to alkylation-induced colon cancer initiated by the alkylating agent, azoxymethane (in combination with the inflammatory agent, DSS), indicating that loss of AAG can protect against toxicity but not necessarily carcinogenesis55, 56 (TABLE 2). Moreover, it is unknown why only some cells and tissues are sensitive to AAG-mediated alkylation toxicity. For example, the MMS-mediated retinal degeneration observed in mice only occurs in the photoreceptor cells of the outer nuclear layer but not in the adjacent layers of the retina30. While it is possible that different tissues might receive different doses of an alkylating agent leading to differential toxicity, most of the aforementioned alkylating agents act directly, without the need for metabolic activation. Instead, it is more likely that insufficient activity of downstream BER enzymes in the sensitive cell types causes an accumulation of toxic BER intermediates that triggers the majority of AAG-dependent cell death in wild-type and Aag Tg mice.
Imbalancing the BER pathway at points downstream of the initiating DNA glycosylase can also sensitize cells to alkylating agents57. For example, if AP endonuclease (APE) is limiting for the repair of AP sites (either because glycosylase levels are too high or APE levels are too low), the consequence is an accumulation of mutagenic and cytotoxic AP sites and the subsequent generation of mutations and strand breaks during replication34, 58, 59 (FIG. 5). Additionally, inducing BER imbalance through APE inhibition (either by RNA interference or chemical inhibitors) produces similar deleterious results by increasing the number of toxic AP sites and enhancing alkylating agent-sensitivity in numerous cell types57, 60-63. Likewise, the inhibition of Pol β can also imbalance the BER pathway leading to severe biological consequences. For example, MEFs lacking Pol β are normal in viability and growth characteristics but exhibit exquisite sensitivity to alkylating agents64. Even partial inhibition of Pol β modulates sensitivity to alkylating agents; a 50% reduction in Pol β, as observed in Polb+/− mice, results in increased DNA single-strand breaks, chromosome aberrations and mutagenicity as compared with wild-type mice65. Importantly, loss of AAG expression in Polb-deficient cells can rescue their sensitivity to alkylating agents, illustrating that the MMS-mediated hypersensitivity and mutagenesis in Polb-deficient cells is AAG-dependent. The ability of double mutant MEFs (lacking both AAG and Pol β) to survive alkylating agent exposure indicates that 3meA and other AAG substrates can be bypassed and tolerated by these cells during DNA replication, presumably by the TLS pathway10, 24, 66, 67 (FIG. 5). However, if AAG excises 3meA to initiate BER, then downstream BER enzymes such as Pol β are required to complete BER to prevent an accumulation of toxic BER intermediates31, 68.
In addition to being a DNA polymerase, Pol β possesses an intrinsic DNA lyase activity that excises 5′dRP residues in DNA (FIG. 5). Mutational analysis has revealed that the 5′dRP lyase activity of Pol β is also required for alkylation resistance, since disabling just the 5′dRP lyase function of Pol β is sufficient to confer alkylation sensitivity69. This finding is notable because it confirms that the 5′dRP species is a toxic BER intermediate. It also demonstrates that, unlike the polymerase function of Pol β, there is no redundant pathway for the 5′dRP removal by Pol β, at least in MEFs. Interestingly, another variant of Pol β that is expressed in some human cancers (termed Pol βΔ), is known to have a dominant negative function that essentially inhibits BER70. By inhibiting BER, Pol βΔ-expressing cells exhibit increased sensitivity to the alkylating agent, methylnitrosourea (MNU), and PolbΔ transgenic mice exhibit greater susceptibility to MNU-induced mammary tumorigenesis compared with wild-type mice70. To further underscore the importance of balance within the BER pathway, increased Pol β activity can also result in deleterious consequences since an increased rate of spontaneous frameshift mutagenesis and microsatellite instability is observed following increased Pol β expression71, 72. Thus, perturbation of either Pol β activity or function in the cell can cause a BER imbalance leading to increased cancer susceptibility and enhanced alkylation sensitivity.
As noted above, the XRCC1 scaffold protein plays a major role in coordinating BER by interacting with the aforementioned DNA processing enzymes at sites of alkylation damage73. Not surprisingly, repression of XRCC1 results in BER imbalance and alkylation sensitivity. For example, Xrcc1-deficient cells display impaired BER and accumulate SSBs after alkylating agent treatment74. Consequently, Xrcc1-deficient cells exhibit severe hypersensitivity to numerous alkylating agents as well as increased genomic instability75, 76. Of note, human monocytes lack detectable levels of XRCC1 and are hypersensitive to alkylating agents77, indicating that specific human cell populations can be deficient in BER. Collectively, these data demonstrate that the initiation of the BER pathway by alkylating agents results in an accumulation of toxic and mutagenic BER intermediates that, if uncontrolled, can result in cell death.
The alteration of various BER proteins in cancer patients emphasizes the possibility that imbalanced BER may play a role in cancer etiology (TABLE 1). Indeed, certain polymorphisms in APE as well as alterations in APE expression or subcellular localization can either be a risk factor or prognostic indicator for numerous human cancers78-83. Further, alterations in Pol β are observed in as many as 30% of human cancers84 while polymorphisms in XRCC1 are linked to increased risk of numerous types of cancer85 (TABLE 1). Thus, a more detailed understanding of how the generation of toxic BER intermediates is regulated in different cell types, both normal and cancerous, will surely contribute to improved cancer chemotherapy with alkylating agents.
Chemical modulation of BER for improved chemotherapy
Inhibitors of BER are being vigorously explored as a mechanism to potentiate the response of cancer cells to alkylating agents. Several inhibitors of APE activity have been identified that sensitize a variety of human cell types to alkylating agents; methoxyamine is one such APE inhibitor that is currently being used in the clinic86-88 (reviewed in 32). Methoxyamine blocks BER by reacting with an aldehyde-sugar group of the AP site, causing a stable methoxyamine-AP-intermediate adduct that blocks the endonuclease activity of APE. By inhibiting the proper completion of BER, methoxyamine has been shown to potentiate the cytotoxicity of numerous alkylating agents in a variety of cancer cell types and tumor xenograft models32, 62, 89. In addition to altering the APE substrate to prevent DNA incision, direct inhibition of APE could also achieve the same end point of blocking BER. However, given that APE also has an important role in maintaining certain transcription factors, including nuclear factor-κB and p53 in an active reduced state, care must be taken to inhibit solely the DNA repair function of APE to prevent off-target effects90, 91.
Small-molecule inhibitors of Pol β represent another strategy to effectively imbalance the BER pathway92. As expected from the characterization of Polb-deficient cells, Pol β inhibitors can increase cellular sensitivity to a variety of alkylating agents93. More recently, highly specific inhibitors of Pol β have been developed that dramatically potentiate temozolomide-sensitivity in colon cancer cells94, 95. Further synergism was observed upon cotreatment with both Pol β inhibitors and temozolomide in cells lacking the homologous recombination factor BRCA2, underscoring the importance of multiple DNA repair pathways in repairing alkylation damage96.
Finally, chemical modulation of another DNA repair enzyme, poly(ADP-ribose) polymerase (PARP) has emerged as a potential chemotherapeutic strategy to increase cellular sensitivity to alkylating agents (reviewed in97, 98). PARP plays an important role in the proper repair of SSBs generated during BER by signaling the presence of SSBs to downstream enzymatic repair machinery. The binding of PARP to SSBs leads to PARP activation and to the synthesis of poly(ADP-ribose) chains that facilitate the recruitment of XRCC1, Pol β and ligase to the DNA strand break. Importantly, Parp1-/- mice and Parp1-/- cells exhibit increased sensitivity to various alkylating agents99, 100, indicating that PARP inhibition can serve as an effective combination therapy with monofunctional alkylating agents. Moreover, the discovery of synthetic lethality in BRCA1 or BRCA2 deficient tumor cells upon treatment with PARP inhibitors has strengthened interest in these inhibitors for single agent therapies101, 102. However, it should be noted that similar to the consequences of altering AAG activity, PARP inhibition can result in dramatically different phenotypic outcomes depending upon the cellular context. For example, PARP inhibition in cells with imbalanced BER (due to altered AAG and Pol β activities) results in resistance to alkylating agents, rather than the alkylation sensitivity observed in other cellular contexts103. Thus, it will be clinically important to investigate overall BER function in tumors and patients before using PARP inhibitors104.
Protecting against highly mutagenic and toxic O6meG lesions
As mentioned above, the O6meG lesion is the predominant O-methyl adduct but contributes only ~5% of the total lesions induced by methylating agents, compared to the 60-80% represented by N-methyl adducts. However, depending on the particular cell type and an organism's genetic background, the O6meG lesion can have dramatic biological effects by eliciting most of the mutagenic and cytotoxic effects associated with SN1 chemotherapeutic alkylating drugs. Similar to the complex biological response caused by 3meA adducts, it is the interplay between numerous factors in several different pathways that determine the final biological outcome in response to O6meG.
The MGMT (alkyltransferase) DNA repair protein plays a pivotal role in governing the fate of organisms after exposure to chemotherapeutic alkylating agents by directly reversing O6meG lesions in genomic DNA17, 105 (FIG. 3 and and4).4). The protective effect of MGMT has been demonstrated in striking fashion using transgenic mice that overexpress either the human or bacterial homologue of MGMT in the thymus, liver or colon; these MGMT-overexpressing mice display a significant reduction in alkylation-induced thymic lymphoma, liver tumor development and colon carcinogenesis106-111 (TABLE 2). Increased expression of MGMT in skin keratinocytes can also confer protection against epidermal papilloma and tumor formation induced by topical application of alkylating agents112-114. Even in cancer-prone mouse models, increasing the level of MGMT activity is sufficient to decrease spontaneous hepatocellular carcinoma as well as alkylation-induced lymphoma development110, 115-118. Remarkably, MGMT-overexpressing mice display significant preservation of cerebellar development and motor function after treatment with alkylating agents, indicating that MGMT can protect against alkylating agent induced toxicity in the brain as well119. Thus, unlike AAG overexpression that sensitizes many cell types to alkylating agents by producing a BER imbalance, the overexpression of MGMT increases cellular resistance to alkylating agents by increasing the amount of direct repair activity in the cell.
Based upon the protective effects of MGMT expression, it is not surprising that loss of MGMT expression can adversely affect survival upon exposure to alkylating agents. Indeed, Mgmt-/- mice exhibit increased levels of cell death in rapidly proliferating tissues such as bone marrow, intestine, thymus and spleen after treatment with alkylating agents (TABLE 2). The tremendous loss of leukocytes and platelets in the haematopoietic stem cell compartment leads to significant ablation of myeloid and lymphoid tissue in Mgmt-/- mice treated with alkylating agents120-122. Notably, the severe pancytopenia that develops in Mgmt-/- mice after alkylation treatment is lethal and can only be rescued by bone marrow transplantation123. Neuronal cell development in the cerebellum and motor function are also severely disturbed in Mgmt-/- mice after treatment with alkylating agents, consistent with a protective function of MGMT in the brain119. In addition to severe tissue loss, alkylation treatment of Mgmt-/- mice results in a large number of thymic lymphomas as well as colon carcinomas that are not detected in wild-type mice treated with the same dose of alkylating agents55, 124-126.
Notably, many types of tumors, including brain, breast, colon and lung, display increased MGMT activity when compared to the corresponding normal tissue (reviewed in 127). While the relation between MGMT activity and clinical outcome remains to be determined for many cancer models, a significant inverse correlation between MGMT expression and patient response has been demonstrated for several types of brain tumors (TABLE 1). In particular, numerous studies have found that pediatric brain tumors exhibit much higher MGMT activity than adults tumors128, 129, leading to a poor response of pediatric tumors to alkylating agents such as temozolomide. Moreover, epigenetic silencing of MGMT expression correlates with a better therapeutic response in patients with glioblastoma130, consistent with the absence or low expression of MGMT in promoter hypermethylated tumors conferring sensitivity to SN1 alkylating agents131.
Imbalancing MGMT activity to improve chemotherapy
The dramatic effect of MGMT activity on cellular sensitivity and resistance to alkylating agents suggests that modulation of MGMT levels could be used to enhance chemotherapy with alkylating drugs. For example, the inhibition of MGMT activity could be used in the clinic to sensitize cancer cells to alkylating agent-induced toxicity. Indeed, several chemical inhibitors of MGMT activity have been developed and shown to improve the efficacy of alkylating agent-based chemotherapy105, 132. Many of these MGMT inhibitors are O6-guanine derivatives that take advantage of the self-inactivating DNA repair reaction of MGMT in which the MGMT enzyme is rendered inert after it catalyzes transfer of an aberrant O6-methyl group onto the active site cysteine in itself. The O6-guanine derivatives act as substrate analogues to bona fide O6meG targets in DNA by reacting with and inactivating endogenous MGMT enzymes. One of the most potent inhibitors in this class is O6-benzylguanine (O6-BG), which has been shown in numerous studies both in vitro and in vivo to rapidly inactivate MGMT activity. Preclinical studies with a wide range of human tumor xenografts have clearly demonstrated the effectiveness of O6-BG in sensitizing cancer cells to SN1-alkylating agents such as BCNU and temozolomide (reviewed in132). Based on its effectiveness, a combination therapy of O6-BG with BCNU has shown some promise in phase II clinical trials treating patients with recurrent glioblastoma133, 134. Unfortunately, MGMT inhibition in non-tumor cells can lead to significant myelosuppression, a complication that must be rectified to specifically target cancer tissues for sensitization to alkylating agents135, 136.
As a corollary to decreasing MGMT activity in cancer cells, increasing MGMT activity in noncancer cells could improve chemotherapeutic efficacy by protecting normal, bystander cells from alkylating agent-induced cell death and mutation. For example, increasing MGMT activity in mouse bone marrow cells can reduce the myelosuppression associated with alkylating agent treatment, suggesting a possible approach for protecting cancer patients from chemotherapy-associated bone marrow toxicity137-140. Of clinical relevance, enhancing MGMT repair activity in haematopoietic cells by stable MGMT overexpression has proven to increase cellular survival during a clinical chemotherapy regimen by mitigating the toxic effects of O6meG adducts141. Chemoprotection has been further demonstrated using immunodeficient mice engrafted with human haematopoietic stem cells overexpressing MGMT, as well as with large animal canine models142-146, suggesting feasibility in human cancer patients. In addition, increased MGMT expression in bone marrow cells would also be expected to reduce the incidence of therapy-related secondary cancers. Ideally, one would express an O6-BG-resistant form of MGMT in the haematopoietic cells of patients undergoing combination chemotherapy with a SN1 alkylating agent plus O6-BG. Such O6-BG-resistant MGMT variants have been extensively characterized and could prove efficacious in future studies132.
Multiple cellular pathways determine the biological effects of O6meG lesions
Although MGMT is a major protective factor against the deleterious consequences of O-alkylation DNA damage, the relative levels of MGMT in a particular tissue are not necessarily predictive of its sensitivity to alkylating agents when MGMT is absent from that tissue. For example, wild-type bone marrow cells express extremely low levels of MGMT (~100-fold less than liver cells) but they are one of the most sensitive tissues in Mgmt-/- mice upon alkylating agent treatment121. Moreover, cells deficient in MGMT can acquire resistance to alkylating agents that is MGMT-independent. These observations indicate that additional cellular mechanisms play significant roles in the response to alkylation damage that must be taken into consideration when accessing the relative sensitivity of a particular cell or tissue type.
In particular, a number of DNA damage repair and bypass pathways converge on O6meG lesions to elicit a variety of downstream effects that can greatly influence the final response to alkylating agents (FIG. 3 and and6).6). The recruitment of multiple cellular pathways to sites of O6meG adducts can be attributed to the DNA replication blocking and miscoding properties of O-methyl lesions, which can inhibit DNA synthesis but ultimately generate DNA mispairs. In the event of replication blocks caused by O6meG, the TLS pathway could provide a cellular mechanism to bypass stalled DNA replication forks. Indeed, it has been shown that TLS polymerases such as Pol η, κ and ζ can bypass O6meG lesions on DNA templates and that genetic depletion of certain TLS polymerases affects cellular sensitivity to SN1 alkylating agents24, 147-152. Unfortunately, due to the miscoding properties of O6meG adducts, replication past these lesions by either TLS or conventional DNA polymerases results in increased mutagenesis through the generation of O6meG:thymine (O6meG:T) mispairs153, 154 (FIG. 6). As described below, the formation of O6meG:T mispairs has dramatic biological consequences by recruiting the MMR pathway that in turn triggers a cascade of DNA processing events that can lead to replication fork collapse, SCEs, chromosome aberrations and cell death.
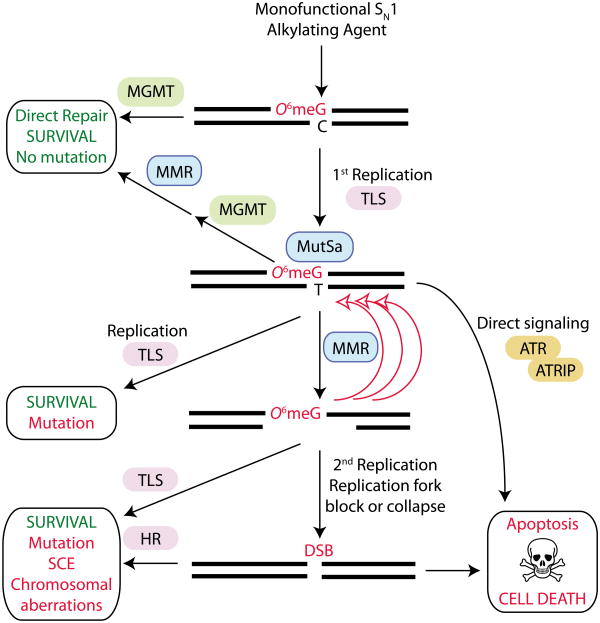
SN1 alkylating agents induce the formation of O6-methylguanine (O6meG) lesions in DNA, which are directly repaired by O6-methylguanine methyltransferase (MGMT). If the O6meG lesion is not repaired, DNA replication by either conventional or translesion synthesis (TLS) polymerases can lead to the formation of O6meG:thymine (O6meG:T) mispairs. Recognition of O6meG:T by the MUTSα protein complex initiates the mismatch excision repair (MMR) pathway leading to futile cycles of DNA resection and processing that can cause replication fork collapse and double strand break (DSB) formation. The cytotoxic DSB can be repaired by the homologous recombination (HR) or non-homologous end-joining (NHEJ) pathway resulting in cell survival but with mutation or sister chromatid exchange (SCE). If the DSB is not repaired, it can trigger programmed cell death by apoptosis. Alternatively, MMR-dependent recognition of O6meG:T can directly signal for programmed cell death through the ATR (ataxia-telangiectasia mutated- and Rad3-related)-ATRIP (ATR-interacting protein) signaling pathway.
Among the cellular factors governing sensitivity to chemotherapeutic alkylating agents, the MMR pathway has emerged as a key determinant of the biological effects of O-alkylation damage (reviewed in155). Although the mechanism by which MMR combines with O-alkyl lesions to modulate the cytotoxicity of alkylating agents is not completely understood, numerous studies have established that recognition of O6meG:T mispairs by the MutSα subunit (comprised of MSH2 and MSH6) of the MMR pathway constitutes a critical step for the initiation of programmed cell death in response to alkylation damage156-160. A possible mechanism to explain the role of MMR in alkylating agent induced cell death involves the repeated processing and regeneration of O6meG:T mispairs, ultimately leading to cytotoxic DNA DSBs (FIG. 6). In this model, the MMR machinery would excise the newly synthesized DNA strand containing the mismatched thymine, but then reinsert another thymine across the O6meG leading to “futile” cycles of MMR. This constant MMR-dependent processing of DNA at O6meG:thymine mispairs would promote the formation of DNA strand gaps161 that can collapse DNA replication forks leading to the formation of DSBs162. The conversion of O6meG lesions into highly cytotoxic DNA DSBs is thought to be the ultimate trigger for cell death by the apoptotic pathway.
In response to collapsed replication forks or DSBs caused by aberrant MMR-dependent processing, error-free recombination via the HR pathway can provide an additional cellular survival mechanism against the cytotoxic effects of O-alkyl lesions163-165 (FIG. 6). Consistent with the formation of DSBs caused by MMR-dependent processing and subsequent repair by HR, cells that are deficient in the HR pathway display increased sensitivity to SN1 alkylating agents166-168. While the HR pathway promotes survival by resolving DNA repair intermediates or DSBs caused by MMR processing, this pathway increases the amount of recombination and SCEs153. Indeed, MGMT-deficient cells treated with non-toxic doses of SN1-alkylating agents display SCEs after two rounds of replication169, consistent with recombination occurring at DNA strand breaks induced by aberrant MMR processing. Thus, in addition to MGMT, at least three different DNA damage repair or bypass pathways (MMR, TLS and HR) can converge at sites of O-alkylation damage to influence the final biological effects caused by a single type of damaged DNA base lesion (FIG. 6).
In addition to MMR-induced DSB formation, the recognition of O6meG:T mispairs by MMR proteins could serve, in and of itself, as a sensor for DNA alkylation damage that directly signals for cell death via apoptosis (FIG. 6). In support of MMR-dependent DNA damage signaling, the MutSα complex can physically interact with and activate the DNA damage checkpoint kinase, ataxia-telangiectasia and Rad3-related (ATR), after the formation of O6meG:T mispairs170. While it is not known whether ATR activation by MMR is sufficient to trigger apoptosis, it is possible that a combination of MMR signaling and MMR-dependent processing of O6meG lesions contribute to the activation of apoptosis. Importantly, in either scenario, alkylation-induced cell death is dependent on the MMR-dependent recognition of O6meG lesions. Thus, in contrast to MGMT, where inhibition or loss of expression increases cellular sensitivity to chemotherapeutic alkylating agents, loss of the MMR pathway produces cells that ‘tolerate’ O-alkyl adducts and are remarkably resistant to the killing effects of SN1-alkylating agents160, 171-175. The protective effect conferred by loss of the MMR pathway has been observed in many different cell types as well as in tissues such as the intestine, colon and haematopoietic stem cell compartment176-181. Notably, the loss of MMR can completely rescue the alkylation-sensitivity of Mgmt-/- mice indicating that MMR-dependent recognition of O6meG lesions is the major contributing factor to cell death and that an entire organism can survive with unrepaired O6meG lesions present in its genomic DNA as long as these lesions are not processed by the MMR machinery122, 182. However, as expected by the mutagenic properties of O-alkylation damage, mice deficient in both MMR and MGMT are extremely sensitive to alkylation-induced lymphoma, demonstrating that the O6meG adducts that escape MMR processing can generate mutations that lead to cancer177, 181, 182. Collectively, these studies indicate that in certain cellular contexts, O-alkyl lesions are the main contributors to the cytotoxicity and mutagenicity of chemotherapeutic alkylating agents with MGMT, MMR, TLS and HR being the pivotal factors in determining eventual toxicity. Importantly, dysregulation in any of these pathways, either through inactivation or overexpression, have been linked to increased cancer susceptibility or cancer resistance to chemotherapeutic alkylating agents (reviewed in 183-187) (TABLE 1).
Concluding remarks and future perspectives
It is becoming clear that an organism's response to chemotherapeutic alkylating agents is dictated by the coordination of factors within a particular DNA repair pathway, as well as by the interplay between different DNA repair pathways. As discussed above, an imbalance in just a single factor can have severe consequences on the health of an organism when exposed to a chemotherapeutic alkylating agent and in some cases, the actions of DNA repair proteins can be highly toxic to the cell rather than protective. Cellular imbalances can arise from genetic differences (mutations, polymorphisms) or epigenetic variation (heterochromatic silencing, microRNA-mediated transcriptional regulation) that can alter the levels of protein expression or activity within a DNA repair pathway188-191. Not surprisingly, these changes in gene expression and regulation can account for the extensive range of alkylating agent-responses exhibited by different cells and tissues within an individual (intraindividual) as well as the broad differences between individuals (interindividual)192, 193. Thus, the next step in understanding how cells respond to chemotherapeutic alkylating agents will involve systems-level analyses (genomics, transcriptomics, proteomics etc.) to provide a global overview of how the levels of DNA repair components can ultimately affect alkylation sensitivity or resistance. The combination of systems-level approaches will improve personalized chemotherapeutic regimens by pinpointing the threshold response of an individual, thereby reducing toxicity or mutagenicity while maximizing tumor cell killing efficiency104.
In addition to the known DNA repair pathways described above, toxicogenomic studies have revealed novel cellular factors that play major roles in determining an organism's response to alkylating agents193-195. The identification of pathways outside of DNA repair that modulate the intra- and interindividual responses to alkylating agents will be of paramount importance for the development of novel chemotherapy regimens that target non-DNA repair pathways with the potential of reducing toxicity and carcinogenicity. Further investigation into the cellular factors and pathways that participate in the repair and response to alkylating agents will provide key insight into the proper use of chemotherapeutic drugs based on alkylating agents. Moreover, these cellular factors and pathways could be altered in normal or cancer cells to improve chemotherapies based on alkylating agents.
Glossary terms
| Alkyl | Chemical sidechain that consists only of single-bonded carbon and hydrogen atoms, for example a methyl or ethyl group. |
| Nucleophilic substitution | Chemical bonding reaction between an electron pair donor nucleophile with an electron pair acceptor electrophile. |
| Depurination | Loss of a purine base (adenine or guanine) from the DNA backbone through chemical or enzymatic hydrolysis. |
| Clastogenic | The ability to disrupt or break chromosomes. |
| Chloroethyl | Alkyl functional group consisting of a chlorine atom bonded to an ethyl carbon group. |
| Sister chromatid exchanges (SCEs) | Crossing over event between sister chromatids leading to the exchange of homologous stretches of DNA sequence. |
| Microsatellite instability | Mutations in short motifs of tandemly repeated nucleotides resulting from replication slippage and deficient mismatch repair. |
| Pancytopenia | Severe reduction in the number of all blood cell types, including red and white blood cells and platelets. |
| Myelosuppression | Inhibition of blood cell production in the bone marrow. |
| Apoptosis | A type of caspase-dependent programmed cell death characterized by cell blebbing and DNA fragmentation. |
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrc3185
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3586545?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nrc3185
Article citations
Bendamustine-rituximab elicits dual tumoricidal and immunomodulatory responses via cGAS-STING activation in diffuse large B-cell lymphoma.
J Immunother Cancer, 12(11):e009212, 09 Nov 2024
Cited by: 0 articles | PMID: 39521616 | PMCID: PMC11551994
Causes and consequences of DNA double-stranded breaks in cardiovascular disease.
Mol Cell Biochem, 15 Oct 2024
Cited by: 0 articles | PMID: 39404936
Review
Molecular Profile as an Outcome Predictor in Glioblastoma along with MRI Features and Surgical Resection: A Scoping Review.
Int J Mol Sci, 25(17):9714, 08 Sep 2024
Cited by: 0 articles | PMID: 39273661 | PMCID: PMC11395592
Review Free full text in Europe PMC
Recognition and Sequencing of Mutagenic DNA Adduct at Single-Base Resolution Through Unnatural Base Pair.
Adv Sci (Weinh), 11(40):e2404622, 03 Sep 2024
Cited by: 1 article | PMID: 39225557 | PMCID: PMC11515917
Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes.
Biomedicines, 12(8):1766, 05 Aug 2024
Cited by: 0 articles | PMID: 39200230 | PMCID: PMC11351110
Go to all (484) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
DNA alkylation lesion repair: outcomes and implications in cancer chemotherapy.
J Zhejiang Univ Sci B, 22(1):47-62, 01 Jan 2021
Cited by: 13 articles | PMID: 33448187 | PMCID: PMC7818015
Review Free full text in Europe PMC
MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents.
DNA Repair (Amst), 6(8):1079-1099, 07 May 2007
Cited by: 369 articles | PMID: 17485253
Review
Alkylation damage in DNA and RNA--repair mechanisms and medical significance.
DNA Repair (Amst), 3(11):1389-1407, 01 Nov 2004
Cited by: 339 articles | PMID: 15380096
Review
The human cyclin B1 protein modulates sensitivity of DNA mismatch repair deficient prostate cancer cell lines to alkylating agents.
Exp Cell Res, 257(1):127-134, 01 May 2000
Cited by: 4 articles | PMID: 10854060
Funding
Funders who supported this work.
NCI NIH HHS (7)
Grant ID: CA112967
Grant ID: CA149261
Grant ID: R01 CA055042
Grant ID: CA075576
Grant ID: R01 CA149261
Grant ID: CA055042
Grant ID: R01 CA075576
NIEHS NIH HHS (3)
Grant ID: DP1 ES022576
Grant ID: P30 ES002109
Grant ID: ES002109
NIH HHS (2)
Grant ID: DP1-OD006422
Grant ID: DP1 OD006422





