Abstract
Free full text

Naturally Occurring Variants of the Dysglycemic Peptide Pancreastatin
Abstract
Pancreastatin (PST), a chromogranin A-derived peptide, is a potent physiological inhibitor of glucose-induced insulin secretion. PST also triggers glycogenolysis in liver and reduces glucose uptake in adipocytes and hepatocytes. Here, we probed for genetic variations in PST sequence and identified two variants within its functionally important carboxyl terminus domain: E287K and G297S. To understand functional implications of these amino acid substitutions, we tested the effects of wild-type (PST-WT), PST-287K, and PST-297S peptides on various cellular processes/events. The rank order of efficacy to inhibit insulin-stimulated glucose uptake was: PST-297S > PST-287K > PST-WT. The PST peptides also displayed the same order of efficacy for enhancing intracellular nitric oxide and Ca2+ levels in various cell types. In addition, PST peptides activated gluconeogenic genes in the following order: PST-297S ≈ PST-287K > PST-WT. Consistent with these in vitro results, the common PST variant allele Ser-297 was associated with significantly higher (by ~17 mg/dl, as compared with the wild-type Gly-297 allele) plasma glucose level in our study population (n = 410). Molecular modeling and molecular dynamics simulations predicted the following rank order of α-helical content: PST-297S > PST-287K > PST-WT. Corroboratively, circular dichroism analysis of PST peptides revealed significant differences in global structures (e.g. the order of propensity to form α-helix was: PST-297S ≈ PST-287K > PST-WT). This study provides a molecular basis for enhanced potencies/efficacies of human PST variants (likely to occur in ~300 million people worldwide) and has quantitative implications for inter-individual variations in glucose/insulin homeostasis.
Introduction
Pancreastatin (PST)3 is a 52-mer endogenous dysglycemic peptide (human sequence: GESRSEALAVDGAGKPGAEEAQDPEGKGEQEHSQQKEEEEEMAVVPQGLFRG-NH2) (1, 2). It was first detected in porcine pancreas and named after its first described effect, i.e. inhibition of glucose-induced insulin secretion from pancreatic islet β cells (3, 4). PST is processed from the pro-hormone chromogranin A (CHGA), a major protein in the endocrine, neuroendocrine, and neuronal tissues by different classes of proteases (such as prohormone convertases, cathepsin L, and trypsin) along with several other bioactive peptides (e.g. catecholamine-release inhibitory peptide catestatin, vasodilatory peptide vasostatin, antimicrobial peptides pro-chromacin/chromacin, and myocardial β-adrenergic-like agonist serpinin (1, 5,–7). PST has been reported to exert diverse biological effects (reviewed in Ref. 1). It inhibits insulin secretion stimulated by physiological activators (e.g. glucose and glucagon) as well as pharmacological agents (e.g. sulfonylurea). PST also inhibits insulin-stimulated glucose uptake and translocation of GLUT4 in primary rat adipocytes (8). In humans, infusion of PST through brachial artery decreases forearm glucose uptake by skeletal muscle (9). PST has also been reported to regulate both carbohydrate and lipid metabolism in humans (10). In addition, PST increases hepatic gluconeogenesis and suppresses insulin signaling via a conventional protein kinase C pathway as well as by the NO pathway (11). PST also inhibits leptin expression and enhances uncoupling protein-2 expression and activates protein synthesis signaling cascade in isolated rat adipocytes (12, 13). Plasma PST levels are elevated in type 2 diabetic patients (10). Consistent with these observations, mice lacking PST (generated by targeted ablation of the CHGA gene (14)) display reduced insulin resistance (apart from elevated systolic and diastolic blood pressure) and exogenous administration of human PST peptide restores insulin sensitivity in knock-out mice (15). Thus, PST is emerging as an important regulator for a number of metabolic functions, including physiological homeostasis of glucose and insulin.
Re-sequencing of the CHGA locus in a Southern California population (n = 180) led to the identification of three non-synonymous variants in the PST domain: R253W, A256G, and G297S (16). Among these naturally occurring PST variants, G297S is located within the functionally active C terminus of the peptide. Interestingly, the G297S variant exhibited ~6-fold stronger inhibition of insulin-stimulated glucose uptake in primary rat adipocytes as compared with the wild-type (10). However, the status of functional genetic variants of PST in ethnically/geographically different human populations has not been reported. Moreover, the molecular basis of increased potency for the Ser-297 peptide (PST-297S) over the wild-type peptide (PST-WT) to diminish cellular glucose uptake remains unclear.
In this study, we aimed to discover naturally occurring variants in PST in an Indian population (n = 410) by systematic re-sequencing and carry out comparative functional analysis of PST peptides. Three variants (viz. R253W, E287K, and G297S) were detected: whereas the A256G variant (mentioned above; (16)) was absent, the E287K was a novel variant in our study population. Approximately 14% of our study subjects had one or another of these PST amino acid variants. Both Lys-287 peptide (PST-287K) and the PST-297S peptide displayed higher potencies (than PST-WT) to various cellular processes/events, including inhibition of insulin-induced glucose uptake, enhancement of intracellular nitric oxide and Ca2+ levels, and activation of gluconeogenic gene transcription. Consistently, the PST-297Ser allele predicted markedly higher plasma glucose levels (as compared with the PST-297Gly allele) in our study subjects. Additionally, we derived the structures of wild-type and variant PST peptides by molecular modeling and molecular dynamics simulations (because no structural information about PST exists in the literature). Interestingly, the PST-297S and PST-287K peptides showed higher helical content (in corroboration with circular dichroism spectroscopic analysis) than the PST-WT peptide, suggesting that the gain of potency for these PST variant peptides may be due to their more ordered secondary/tertiary structures than the wild-type peptide.
EXPERIMENTAL PROCEDURES
Recruitment of Human Subjects and Discovery of Genetic Variations in the PST Domain
Unrelated volunteers (n = 410) from an urban Chennai cosmopolitan population were recruited at the Madras Medical Mission hospital. Each subject gave informed consent for use of their DNA for genetic analysis. The mean age of the subjects was ~40 years; ~13% of the subjects were diabetics and ~38% subjects were hypertensives; none of the subjects had a history of kidney disease or any type of cancer. This study was approved by the Institute Ethics Committee at Indian Institute of Technology Madras, Chennai. Genomic DNA was isolated from EDTA anti-coagulated blood samples using the FlexiGene DNA kit (Qiagen). Exon 7 of CHGA was PCR amplified using PhusionTM high-fidelity DNA polymerase (New England Biolabs) and the following primers: forward, 5′-GAGTGGCAGAGACTGGGAAAATG-3′; reverse, 5′-ACAGAGCTGGCTCCCGCCC-3′. The detailed PCR protocol has been reported recently (17). The PCR products were purified by using QIAquick PCR Purification columns (Qiagen) and sequenced using an Applied Biosystems 3130 Genetic Analyzer (USA) and the forward primer (as mentioned above). Genetic variations were detected and confirmed from chromatograms. To confirm any ambiguity about a polymorphism re-sequencing of the sample was carried out using the reverse primer (as mentioned above).
Cell Culture and Transfection Conditions
L6, HepG2, and 3T3-L1 cells were grown and maintained in 25-cm2 tissue culture flasks (Nunc, USA) in Dulbecco's modified Eagle's medium (high glucose) containing 100 units/ml of penicillin, 100 μg/ml of streptomycin (Invitrogen), and 10% heat inactivated fetal bovine serum (FBS) in a 5.5% CO2 environment at 37 °C in a humidified incubator (Thermo Scientific). L6 and HepG2 cells were passaged every 3–4 days and 3T3-L1 cells were differentiated as described later.
Synthesis of Human PST Peptides
The human PST peptides were synthesized by solid phase method and purified to at least 95% homogeneity by GenPro Biotech, New Delhi, India. Authenticity and purity of these peptides were verified by analytical high performance liquid chromatography and mass spectrometry. The amino acid sequences of PST peptides synthesized were: wild-type (PST-WT, PEGKGEQEHSQQKEEEEEMAVVPQGLFRG-amide), Ser-297 variant (PST-297S, PEGKGEQEHSQQKEEEEEMAVVPQSLFRG-amide) and Lys-287 variant (PST-287K; PEGKGEQEHSQQKEKEEEMAVVPQGLFRG-amide); the bold and underlined residues indicate the variant positions. The peptides were dissolved in water at 1 mm concentration and stored as single use aliquots at −80 °C.
Measurement of Glucose Uptake by Differentiated 3T3-L1 Adipocytes
3T3-L1 cells were differentiated by growing the cells in the presence of 500 μm isobutylmethylxanthine (Sigma), 25 μm dexamethasone (Sigma), and 4.0 μg/ml of insulin (Sigma) for 3 days and then in the presence of 4.0 μg/ml of insulin for an additional 3 days as described previously (18). The cells were then grown in normal growth media until fully differentiated, typically by 10 days (from the point of seeding). The adipocytes were trypsinized and re-seeded into six-well plates. On the day of glucose uptake assay, cells were maintained in the normal growth medium without serum for 2 h. Next, the cells were incubated in Krebs-Ringer Hepes buffer without glucose in the presence or absence of PST peptides (at 100 nm final concentration) for 10 min before stimulation with 100 nm insulin for 20 min. The cells were then incubated with 0.5 μCi of [3H]2-deoxy-d-glucose (PerkinElmer Life Sciences) (i.e. 0.1 mm 2-deoxy-d-glucose) for an additional 20 min. After incubation, cells were washed with ice-cold phosphate-buffered saline. The cells were finally lysed with 0.5 m NaOH and 0.1% SDS. Radioactivity in the cell lysates was measured in a TricarbTM liquid scintillation counter (PerkinElmer Life Sciences) and the total protein was measured by Bradford reagent.
Measurement of Nitric Oxide (NO) Levels in Cultured Cells
NO levels in L6 cells were measured by the 4,5-diaminofluorescein diacetate method as described previously (19). Briefly, L6 cells were seeded in 24-well plates. At 60–70% confluence (24 h after seeding), cells were serum starved for 6 h and then treated with 10 nm PST-WT or PST-287K or PST-297S in the presence or absence of 200 μm l-NAME (an analog of arginine that inhibits NO production; Sigma) for 15 min. All peptides and l-NAME solutions were prepared in serum-free media. After 15 min of treatment, cells were washed twice with phosphate-buffered saline (PBS) and incubated with 5 μm 4,5-diaminofluorescein diacetate (Sigma) and 1.0 mm l-arginine (nitric-oxide synthases (NOS) substrate; Sigma) in serum-free media for 10 min at 37 °C. The cells were then washed twice with PBS and the fluorescence was detected by a Nikon-Ti Eclipse fluorescence microscope (Japan) with excitation wavelength of 485 nm and emission wavelength of 530 nm. The mean fluorescence intensity was calculated by ImageJ analysis of n = 30 cells/condition.
Estimation of Intracellular Free Ca2+ Levels
Intracellular Ca2+ levels in L6 cells in the presence or absence of PST peptides were estimated by a ratiometric method using Fura 2-AM dye (Invitrogen). In brief, cells were seeded onto coverslips at 60–70% confluence and washed with Krebs-Ringer Hepes buffer (KRB) containing (125 mm NaCl, 5 mm KCl, 1 mm Na2HPO4, 1 mm MgSO4, 1 mm CaCl2, 5.5 mm glucose, and 20 mm HEPES, pH 7.2) or Ca2+-free bath solution (KRB) containing 5 mm EGTA and incubated with 5 μm Fura 2-AM (Invitrogen) at room temperature (22–24 °C) for 30 min. Cells were then washed in the bath solution with or without Ca2+ for 30 min and illuminated with dual-excitation wavelengths (at 340 and 380 nm). Emission was captured at 510 nm with appropriate excitation and emission filters. Images were acquired by an ANDOR CCD camera attached with an Olympus IX71 inverted microscope, controlled through Andor iQ software (ANDOR Technologies). Cells were perfused for 2 min with bath solution with or without Ca2+ before the perfusion of PST peptides (PST-WT, PST-287K, and PST-297S) at 10 nm concentration through channels/injectors. These peptides were perfused continuously for 10 min and then washed with bath solution with Ca2+ or zero Ca2+ to bring down the PST-induced effect to the basal levels. Regions of interest were drawn to define borders of cells and fluorescence ratios (340/380 nm) were calculated from the background subtracted images (n = 15 cells/condition). The rise in intracellular Ca2+ levels (indicated by an increase in the 340/380 nm values) in the presence of each peptide was compared with the corresponding control (i.e. under perfusion with bathing solution with Ca2+ or without Ca2+).
Generation and Expression of Human Pepck-1 and G6Pase Promoter-Luciferase Reporter Constructs
An approximately 1.3-kb segment of human phosphoenolpyruvate carboxykinase 1 (hPepck-1) promoter (from −1317 to +64 bp; numberings are with respect to the transcription initiation site as +1) was PCR-amplified from human genomic DNA using the following primers: forward, 5′-GGAGATCTGGGTTCTCTAAGTGAGTTTGGTCG-3′ (the bold nucleotides indicate an added BglII site) and reverse, 5′-TCGGATCCCGCCAGCAAGTTTGTGTTCC-3′ (the bold nucleotides indicate the added BamHI site). These primers were designed based on the reference sequence for the hPepck-1 gene (NM_002591). The PCR product was purified and restriction-digested with BglII and BamHI and cloned into a promoter-less Gaussia luciferase reporter vector (pGLuc-Basic Vector, New England Biolabs). The promoter sequence in the clone was confirmed by sequencing with pGLuc-FP (5′-GGGGTTCCGCGCACATTTCCCCG-3′) and pGLuc-RP (5′-TCAGGGCAAACAGAACTTTGACTC-3′) primers. The resultant plasmid was denoted as the hPepck-1 construct. Similarly, an approximate 1.3-kb region of the human glucose 6-phosphatase (hG6Pase) promoter (from −1293 bp to +23 bp) was PCR amplified using the following primers: forward, 5′-CCAAGCTTGCCAGGCATGGTGGTTCAC-3′ (the bold nucleotides indicate an added HindIII site) and reverse, 5′-GGGGTACCGCTTGGTGGTGATTGCTCTGC-3′ (the bold nucleotides indicate an added KpnI site). These primers were designed based on the reference sequence for the human hG6Pase gene (NM_000151). The PCR product was cloned between HindIII and KpnI sites of the pGLuc-Basic vector; the correct insertion/orientation of the cloned DNA fragment was confirmed by sequencing. The recombinant plasmid thus obtained was named the hG6Pase construct.
The hPepck-1 or hG6Pase constructs (1.0 μg/well) were transfected into HepG2 cells (at 70–80% confluence) in 12-well plates by Transpass D2 transfection reagent (New England Biolabs) to check its expression pattern in the absence or presence of various concentrations (0.05, 0.1, 0.5, 1.0, 10, and 50 nm) of PST peptides. After 12 h of transfection, cells were treated with one of the three PST peptides in low glucose DMEM F-12 media (Hi-Media, India) and 24 h after treatment, media (supernatant) from each well was collected and assayed for luciferase activity using a BioLux® Gaussia luciferase assay kit (New England Biolabs). The cells were lysed with lysis buffer (100 mm phosphate buffer, pH 7.8, 1.0 mm dithiothreitol, and 0.1% Triton X-100) and total protein was estimated by the Bradford reagent (Bio-Rad). Luciferase values were normalized with the total protein. As a control, the promoter-less pGLuc-Basic vector was transfected in some experiments; under the basal/untreated conditions, hPepck-1 and hG6Pase constructs showed ~25-fold higher activity than the pGLuc-Basic vector. In another set of experiments, the hPepck-1 construct was transfected into L6 cells and those were treated with PST peptides.
Generation of Modeled PST Peptide Structures
Because the PST structure is not available in the Protein Data Bank (PDB) and BLAST against PDB did not yield a suitable template for homology modeling of PST we undertook a protein-fold recognition (threading) approach to develop its structure using the FUGUE server (20). Interestingly, FUGUE recognized the myocyte-specific enhancer factor 2b protein as the closest homologue of PST-WT. Therefore, the PST-WT primary sequence was threaded onto the sequence of this protein (PDB code 1TQE). Next, Modeler 9v10 (21) was employed to generate the three-dimensional coordinates for PST-WT based on the output alignment of FUGUE. Among the 40 generated models, the best model was chosen based on the optimized modeler objective function. Residues Glu-287 and Gly-297 were then mutated to Lys-287 and Ser-297, respectively, using Modeler 9v10 and a short energy minimization was done to re-position the side chains. The final refinements of these structures were done by molecular dynamics (MD) simulations.
All MD simulations were done in GROMACS-4.5.5 software (22), using CHARMM36 force-field parameters (23, 24). Briefly, the systems were further energy minimized for 1000 steps using the conjugate gradient and another 1000 steps using the steepest descent algorithm. After relaxing the peptides in gas phase, the structures were solvated in a cubic periodic box of explicit water with water molecules extending 9 Å outside the peptide on all sides. The 3-site TIP3P model was chosen to describe the water molecules. The salt concentration of 0.15 m was maintained by including appropriate numbers of Na+ and Cl− ions. Subsequently, an extensive set of minimization and thermalization of the engineered structures was performed by gradually heating to 310 K in a canonical ensemble. The solvent density was adjusted under isobaric and isothermal conditions at 1 atm and 310 K. The systems were equilibrated for 10 ns in NPT ensemble, with a simulation time step of 2 fs. These structures were further simulated to generate the 200 ns production data. The two variants were also simulated for 200 ns following the same protocol. The long-range electrostatic interactions were treated by using Particle-Mesh Ewald sum and SHAKE was used to constrain all bonds involving hydrogen atoms. The C-terminal of the peptides was amide-capped to mimic the physiological conditions. MD simulations for all three peptides were also carried out in the presence of 0.15 m NaCl and 50% 2,2,2-trifluoroethanol (TFE) for 200 ns following the same protocol.
Circular Dichroism (CD) Spectroscopic Analysis
CD spectroscopic analysis of peptides was carried out as described by us previously (25) with some modifications. In brief, lyophilized PST peptides (PST-WT, PST-287K, and PST-297S) were dissolved in PBS buffer, pH 7.4, with 0.01% sodium azide to a concentration of 1 mm. For CD spectroscopy measurements, the final peptide concentration was adjusted to 50 μm in PBS. To study TFE-induced secondary structural changes, TFE was added to the stock peptide solution such that the peptide concentration became 50 μm and the final TFE concentrations (v/v) were 50, 60, and 80%. CD measurements were performed using a JASCO spectropolarimeter (model J-810). Spectra were recorded over the wavelength range of 198–260 nm. Relative quantities of random coil, α-helix, β-sheet, and β-turn were determined by spectral deconvolution of CD data using the CDPro software package as described earlier (25).
Data Presentation and Statistical Analysis
The experimental data presented are representative of at least three separate experiments. Results are expressed as mean ± S.E. from replicate cells or wells as indicated in the respective figure legends. Student's t test and one-way ANOVA followed by Tukey's multiple comparison post hoc test, as appropriate, were performed using GraphPad Prism version 5.04. Genotype-phenotype association analyses were carried out for PST-297-Gly/Gly and PST-297-Gly/Ser genotype groups. Genotype-specific associations were determined by a Levene test for equality of variances and t test for equality of means using SPSS software (SPSS Inc., Chicago, IL). Statistical associations between genotypes and phenotypes were also tested separately in males and females. A p value of ≤0.05 was chosen for statistical significance between groups.
RESULTS
Identification of Non-synonymous Genetic Variations in the Human PST Sequence
To probe for genetic variations in the PST peptide that might alter its function we re-sequenced the genomic region of CHGA in 410 individuals (i.e. 820 chromosomes). We discovered three non-synonymous (amino acid replacement) polymorphisms: C9226T (rs 9658662), which causes substitution of Arg (codon: CGG) by Trp (codon: TGG) at the 253-amino acid position of the precursor protein CHGA; G9328A (ss 748775092), which causes substitution of Glu (codon: GAG) by Lys (codon: AAG) at the 287-residue of CHGA; and G9358A (rs 9658664), which causes substitution of Gly (codon: GGC) by Ser (codon: AGC) at the 297-residue of CHGA (Fig. 1). Among these PST variants, whereas R253W and G297S were reported previously in a Southern California population (10) the E287K variant is a novel one. To confirm this substitution, we carried out reverse-strand sequencing (Fig. 1, panel B). The minor allele frequencies for R253W, E287K, and G297S polymorphisms were 0.24, 0.12, and 6.7%, respectively; together, ~14% of the study population contained one of these PST variants (Table 1). The genotype frequencies for each of these variants were in Hardy-Weinberg equilibrium (R253W: χ2 = 0.002, p = 0.96; E287K: χ2 = 0.001, p = 0.98; G297S: χ2 = 2.119, p = 0.146).

Discovery of amino acid variants of pancreastatin. A, R253W variant. Wild-type CC, heterozygous variant CT at 9226 bp are shown. Here, the 1st nucleotide (indicated by *) of the codon CGG (in the wild-type) is altered to TGG (in the variant) altering the amino acid Arg to Trp. B, E287K variant. Wild-type GG and heterozygous variant GA at 9328 bp are shown. Here, the 1st nucleotide (indicated by *) of the codon GAG (in the wild-type) is altered to AAG (in the variant) altering the amino acid Glu to Lys. C, G297S variant. Wild-type GG, heterozygous variant GA, and homozygous variant AA at 9358 bp are shown. Here, the 1st nucleotide (indicated by *) of the codon GGC (in the wild-type) is altered to AGC (in the variant) that would alter Gly to Ser.
TABLE 1
Non-synonymous genetic variations in PST domain in various ethnic/geographical populations
The genetic variants in the PST domain of the CHGA gene in different populations are listed. The G297S variation displayed high frequencies in Indian populations. Results from this study are shown in bold.
| S. No. | SNP (rs# or ss#) | Population (number of subjects) | MAFa | Ref./Source |
|---|---|---|---|---|
| % | ||||
| 1 | R253W (rs9658662) | Asian (n = 44) | 1.80 | ncbi.nlm.nih.gov |
| Hispanic (n = 28) | 1.00 | ncbi.nlm.nih.gov | ||
| ESP_Cohort_Populations (n = 1594) | 0.10 | ncbi.nlm.nih.gov | ||
| European descent population (n = 662) | 0.40 | ncbi.nlm.nih.gov | ||
| South Indian (n = 410) | 0.24 | This study | ||
| 2 | A256G (rs9658663) | African American (n = 57) | 0.90 | ncbi.nlm.nih.gov |
| Hispanic (n = 28) | 1.80 | ncbi.nlm.nih.gov | ||
| Yoruba in Ibadan, Nigeria (n = 59) | 2.50 | HapMap data | ||
| ESP_Cohort_Populations(n = 1677) | 1.20 | ncbi.nlm.nih.gov | ||
| 3 | E274K (rs371137176) | ESP_Cohort_Populations(n = 6490) | Not available | ncbi.nlm.nih.gov |
| 4 | E287K (ss748775092) | South Indian (n = 410) | 0.12 | This study |
| 5 | E288K (rs140089514) | ESP_Cohort_Populations (n = 1419) | 0.0005 | ncbi.nlm.nih.gov |
| 6 | G297S (rs9658664) | ESP_Cohort_Populations (n = 1360) | 0.50 | ncbi.nlm.nih.gov |
| Utah residents with European ancestry (n = 110) | 1.40 | HapMap data | ||
| Han Chinese, Beijing, China (n = 43) | 1.20 | HapMap data | ||
| Luhya, Webuye, Kenya (n = 89) | 0.60 | HapMap data | ||
| Mexican ancestry in Los Angeles, California (n = 49) | 2.00 | HapMap data | ||
| Toscans in Italy (n = 88) | 1.10 | HapMap data | ||
| African ancestry, Southwest USA (n = 49) | 2.00 | HapMap data | ||
| AGI_ASP population (n = 38) | 1.30 | ncbi.nlm.nih.gov | ||
| Gujarati Indians in Houston, Texas (n = 88) | 6.80 | HapMap data | ||
| South Indian (n = 410) | 6.70 | This study | ||
| 7 | R300Q (rs147297806) | ESP_Cohort_Populations(n = 1350) | 0.0005 | ncbi.nlm.nih.gov |
a MAF, minor allele frequency.
Next, we carried out inter-species sequence alignment of the PST domain of CHGA in different mammals (Fig. 2) to check whether the identified genetic variations occurred previously in the course of evolution. The Arg-253 allele was conserved in primates excepting Marmoset (having Leu; codon: CTG); among the non-primates, rodents had Gln (codon: CAG), whereas giant panda, horse, and rhinoceros had Trp-253; thus, the Trp-253 variant (detected in 2 of 410 subjects) appears to be a reversion. The Glu-287 was conserved among primates as well as non-primates excepting only rodents (that had Gly; codon: GGC). Indeed, the Lys-287 allele was a rare variant that occurred in only 1 of 410 subjects. The Gly-297 allele was also conserved among most of the primates and non-primates with few exceptions: Asp (codon: GAT) in marmoset and giant panda, Val (codon: GTG) in cow and yak. However, the Ser-297 allele (that was not detected in any mammals) occurred in 55 of 410 subjects. In general, the carboxyl terminus of PST was shown to be highly conserved among all the mammalian species included in this analysis (Fig. 2); this observation is consistent with previous reports that the active part of the PST peptide is located within the carboxyl terminus 29 residues (3, 26). Thus, two of three discovered PST variants (viz. E287K and G297S) are located within the functionally active motif of the peptide.
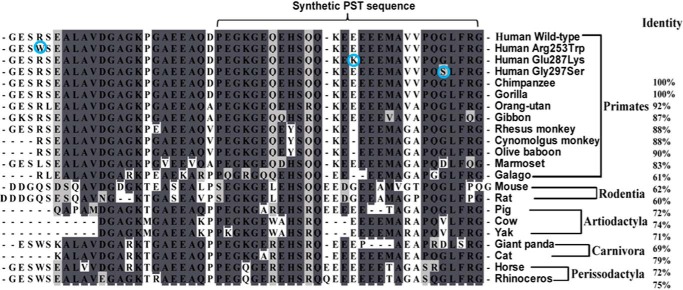
Human pancreastatin variants and inter-species homology among Eutherian mammals. ClustalW multiple sequences alignment was carried out using BioEdit version 7.0.9.0 (Ibis Biosciences, Carlsbad, CA); the percentages of homology are given on the right side. These organisms are arranged according to their Order. Human variants are encircled. The identical amino acid residues from all species are shaded with dark gray and similar amino acid residues are shaded with light gray, and Gap in the alignment is indicated by - - -. All species showed a minimum of 60% similarity in the PST region of CHGA. PST sequences used are: human (accession number NM_001275), common chimpanzee (accession number XP_510135.3), Gorilla (XP_004055643.1), sumatran orangutan (XP_002825091.1), white-cheeked gibbon (XP_003260951.1), rhesus monkey (XP_001092629.2), cynomolgus monkey (BAE01874.1), olive baboon (XP_003902238.1), marmoset (XP_002754260.1); Galago (XP_003787045.1), house mouse (NP_031719.1) and common rat (AEB41037.1); Artiodactyla, wild pig (NP_001157477.2), cow (NP_851348.1), and Yak (ELR47684.1); carnivora: giant panda (XP_002923879.1) and domestic cat (XP_003988016.1); perissodactyla, horse (NP_001075283.1) and white rhinoceros (XP_004434274.1)).
Differential Inhibition of Insulin-stimulated Glucose Uptake by Human PST Peptides
Because alterations within the evolutionarily conserved, active part of human PST (i.e. CHGA-(273–301)) may alter the potency of the peptide, we synthesized Lys-287 and Ser-297-containing peptides (designated as PST-287K and PST-297S, respectively) as well as the wild-type (designated as PST-WT) peptide and tested their effects on insulin-stimulated glucose uptake in differentiated 3T3-L1 adipocytes. Insulin significantly (p < 0.01; compared with the basal condition) augmented 2-deoxyglucose uptake that was blocked by each of the PST peptides (Fig. 3). However, the extents of inhibition of insulin-stimulated glucose uptake were different among the peptides (one-way ANOVA F = 8.16, p = 0.003): PST-297S and PST-287K inhibited ~73 and ~63%, respectively, whereas PST-WT could diminish only ~23%. This observation is consistent with substantially higher potency and efficacy of PST-297S than the PST-WT to inhibit 2-deoxyglucose uptake in rat primary adipocytes (10).
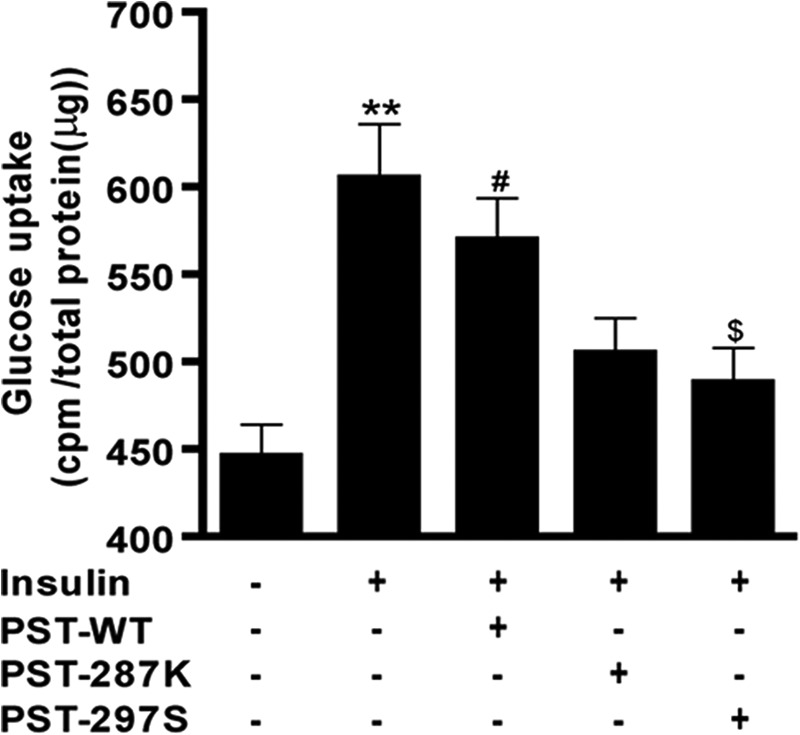
Effect of human PST peptides on insulin-stimulated glucose uptake by differentiated 3T3-L1 pre-adipocytes. Differentiated 3T3-L1 adipocytes were seeded into 6-well plates and each peptide (100 nm) was incubated for 10 min before stimulation with insulin (100 nm). [3H]2-Deoxy-d-glucose (0.1 mm) was also added to the cells after 20 min of insulin treatment. After 20 min of incubation, cells were lysed and radioactivity was measured by scintillation counting. ** represents p < 0.01 for basal versus insulin, # represents p < 0.05 for basal versus insulin + PST-WT, and $ represents p < 0.05 for insulin versus insulin + PST-297S. Each data point represents mean ± S.E. from three separate wells and each experiment was repeated at least three times. The experimental groups are compared by one-way ANOVA followed by Tukey's multiple comparison post test, F = 8.16, p = 0.003. The order of efficacy for the peptides in the inhibition of insulin-stimulated glucose uptake was: PST-297S > PST-287K > PST-WT.
Effect of Human PST Peptides on NO Production
In view of a recent report that PST induces gluconeogenesis by activating the NOS pathway in liver cells (11), we intended to test whether the peptide exerts a similar effect in other cell types and whether the human PST variants differ in their efficacies. Treatment of skeletal muscle L6 cells with PST-WT significantly increased (~164%, p < 0.001) NO production as compared with the basal/untreated condition (Fig. 4). Similarly, PST-287K and PST-297S displayed ~202 (p < 0.001) and ~261% (p < 0.001) higher levels of NO, respectively, as compared with the basal condition, suggesting that both variants are more active than the wild-type peptide (Fig. 4). Interestingly, enhancements in NO levels by each of these PST peptides were completely nullified with the NOS inhibitor l-NAME. Of note, PST peptides exhibited similar effects on NO production in the liver cell line HepG2 (data not shown), suggesting the effectiveness of these peptides across various cell types.

Effect of human PST peptides on nitric oxide production in the L6 cell line. L6 cells cultured in 24-well plates were incubated with l-NAME (200 μm), each of PST peptides (10 nm) and l-NAME together for 15 min. The NO production was observed by incubating cells with 4,5-diaminofluorescein diacetate (5 μm) and l-arginine (1 mm) in serum-free medium for 10 min. Panel A, representative images for the various conditions. Panel B, the fluorescence intensity (NO index) was calculated by ImageJ analysis and plotted as mean ± S.E. *** represents p < 0.001 for basal versus each PST peptide, ### represents p < 0.001 for l-NAME versus each PST peptide, $$$ represents p < 0.001 for PST-WT versus PST-287K, PST-WT versus PST-297S, PST-WT versus PST-WT + l-NAME, €€€ represents p < 0.001 for PST-287K versus PST-297S, PST-287K versus PST-287K + l-NAME, ¥¥¥ represents p < 0.001 for PST-297S versus PST-297S + l-NAME. The experimental groups were compared by one-way ANOVA followed by Tukey's multiple comparison post test, F = 255.7, p = 0.0001, n = 30 cells/condition. The order for efficacy of peptides on NO production was: PST-297S > PST-287K > PST-WT.
Effect of Human PST Variants on Intracellular Free Ca2+ Concentration
Because PST has recently been reported to increase expression of Pepck-1 via conventional protein kinase C (11) and [Ca2+]i is well known to activate conventional PKC, we aimed to test whether human PST variant peptides display differential modulation of [Ca2+]i. Accordingly, L6 cells were pre-loaded with Fura-2 and the fluorescence ratio (340/380 nm) before and after treatment with the human PST peptides was monitored. Each of these human peptides led to a significant rise (over the basal) in the fluorescence ratio values, suggesting increases in [Ca2+]i, in their presence (Fig. 5). The rise in the fluorescence ratio values were different among the peptides (one-way ANOVA F = 32.18, p < 0.0001). Specifically, the PST-297S and PST-287K peptides displayed ~3.8- (p < 0.001) and ~3.1-fold (p < 0.001) increases, respectively, in [Ca2+]i as compared with the PST-WT (Fig. 5, panel D). Interestingly, the PST peptides showed similar effects in evoking [Ca2+]i in L6 cells in the presence of extracellular Ca2+ (Fig. 5, panel E). PST peptides also evoked a similar effect on [Ca2+]i in the pancreatic cell line RINm5F (data not shown), indicating a similar role of these peptides at different cell types/sites of action.
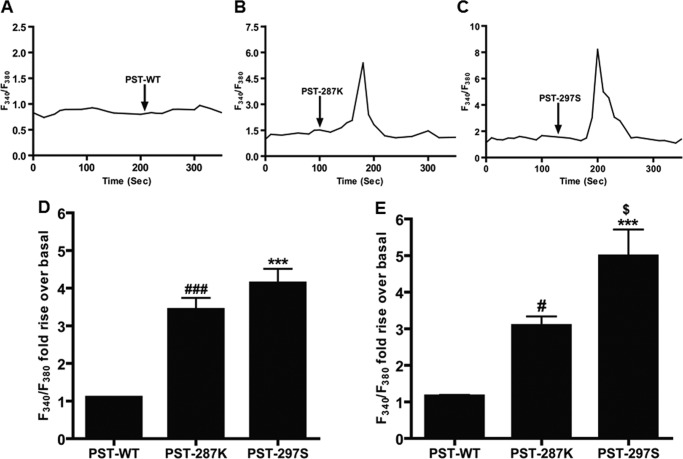
Effect of human PST peptides on intracellular Ca2+ in L6 cell line. L6 cells were seeded onto coverslips and incubated with Fura 2-AM (5 μm) for 30 min. After incubation, cells were washed with Ca2+-free bathing solution for 30 min. After perfusion with bath solution for 2 min, PST peptides (PST-WT, PST-287K, and PST-297S) at 10 nm concentration were perfused to the cells continuously for 10 min. The rise in [Ca2+]i was indirectly measured by dividing the F340/F380 fluorescence ratio from each peptide perfusion and the F340/F380 fluorescence ratio that was observed from the Ca2+-free bath solution perfusion alone. Panels A–C, representative F340/F380 transients in the cases of PST-WT, PST-287K, and PST-297S; arrows indicate the points of addition of PST peptides. Panel D, F340/F380 fold-rise over basal for each PST peptide. ### represents p < 0.001 for PST-WT versus PST-287K and *** represents p < 0.001 for PST-WT versus PST-297S. The experimental groups were compared by one-way ANOVA followed by Tukey's multiple comparison post test; F = 32.18, p < 0.0001, n = 15 cells/condition. Panel E, F340/F380 fold-rise over basal for each PST peptide in the presence of calcium-containing bath solution. # represents p < 0.05 for PST-WT versus PST-287K, *** represents p < 0.001 for PST-WT versus PST-297S, and $ represents p < 0.05 for PST-287K versus PST-297S. The experimental groups were compared by one-way ANOVA followed by Tukey's multiple comparison post-test; F = 18.78, p < 0.0001, n = 10 cells/condition. The order for efficacy of peptides on an increase in [Ca2+]i, both in the absence and presence of extracellular calcium was: PST-297S > PST-287K > PST-WT.
Differential Potencies of PST Variants on Activation of hPepck-1 and hG6Pase Genes
Because PST is known to activate hepatic glycogenolysis (27) and gluconeogenesis in vivo (11), we sought to test whether the human PST peptides differ in their potencies to activate gluconeogenic genes (e.g. hPepck-1 and hG6Pase). Indeed, each of the three peptides activated hPepck-1 and hG6Pase promoter activities in the liver cell line HepG2 (Fig. 6, A and B), consistent with a previous report of induction of endogenous Pepck-1 and G6Pase mRNA levels in mouse liver by PST (11). However, the human PST peptides displayed differential potencies for transcriptional activation of both genes. Specifically, in the case of hPepck-1 promoter, the EC50 values were: PST-297S (0.071 nm) ≈ PST-287K (0.069 nm) < PST-WT (0.098 nm); in the case of hG6Pase, those were: PST-297S (0.01 nm) < PST-287K (0.067 nm) <PST-WT (0.088 nm) (Fig. 6, A and B). The PST-287K and PST-297S peptides also showed higher potencies than PST-WT to augment hPepck-1 promoter activity in L6 cells (Fig. 6C). The variant PST peptides, in general, exhibited higher efficacy and potency than the wild-type PST peptide in activating gluconeogenic genes in liver and skeletal muscle cell types.
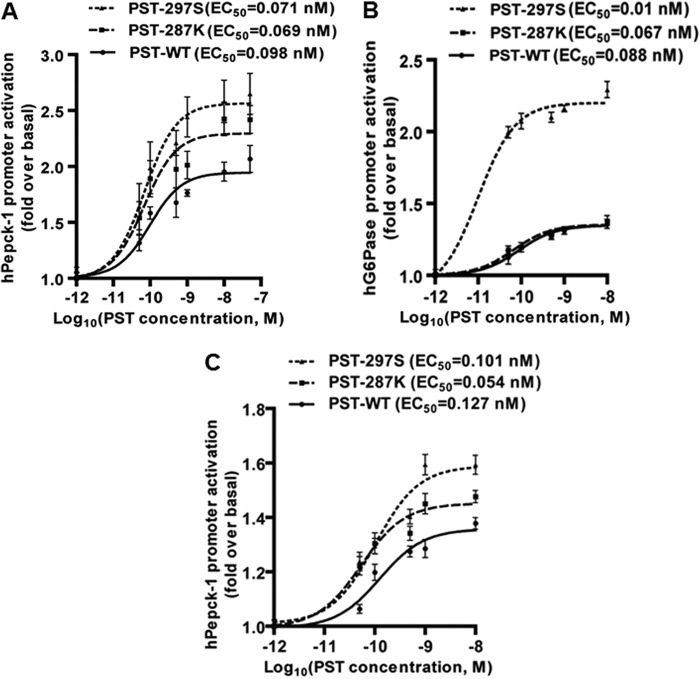
Effect of PST peptides on activation of phosphoenolpyruvate carboxykinase-1 and human glucose-6-phosphatase promoters. HepG2 cells cultured in 12-well plates were transfected with hPepck-1 (panel A) or hG6Pase (panel B) promoter constructs. In another set of experiments, L6 cells cultured in 12-well plates were transfected with the hPepck-1 promoter construct (panel C). After 12 h of transfection, cells were incubated with ascending doses of each PST peptide for 24 h and the luciferase activities in the culture media were determined. Promoter activity is shown as fold-rise with respect to the untreated ones. EC50 values for each peptide were calculated from log(agonist) versus response by 3-parameter non-linear regression analysis. Each data point shows the mean ± S.E. of four separate wells from one representative experiment. The order of efficacy/potency of the PST peptides on hPepck-1 promoter activation, in general, was: PST-297S ≈ PST-287K > PST-WT.
Structures of the Human PST Peptides, Computational Analysis
To derive the possible secondary/tertiary structures of PST-WT and its variants, molecular modeling and molecular dynamics (MD) simulation studies were carried out as crystal or NMR structures of these peptides was not available. An initial model of PST-WT was constructed using FUGUE (20) and MODELLER (21) softwares. FUGUE recognized myocyte-specific enhancer factor 2b protein as the closest homologue, by sequence-structure comparison. The available x-ray crystal structure (2.7-Å resolution) of myocyte-specific enhancer factor 2b was, therefore, used as the template to generate three-dimensional models of PST-WT using MODELLER. Among the 40 models generated, the best model was chosen based on the least number of amino acids in the unfavorable region of the Ramachandran plot; indeed, the best model has no residue in the strictly disallowed region. Next, Glu-287 and Gly-297 residues were mutated to Lys-287 and Ser-297, respectively, in MODELLER to develop the structures of PST-287K and PST-297S peptides. The chosen structures of PST-287K and PST-297S peptides also did not have any residue in the strictly disallowed region of the Ramachandran plot. The PST peptide structures were refined by MD simulations.
The stability of the simulated structures was examined by plotting the time evolution of their root mean square deviations from the starting conformation. Root mean square deviations for all peptides showed an initial increase, but reached to steady mean values in about 25 ns of the simulation time; the PST-287K variant, however, showed large fluctuations at the beginning, but ceased to show a systematic drift after 120 ns (Fig. 7, panel A). The initial drift is expected as the peptide residues reduce any unfavorable interactions and the solvent molecules relax around them. Each of the PST peptides displayed more-or-less similar averaged root mean square deviation values after 120 ns. The subsequent structural analyses of the peptides were, therefore, executed on the last 80 ns data. Fig. 7, panels B–D, shows the evolution of the secondary structural elements of the peptides, as the simulations progressed. It is evident that the peptides acquire more helicity over time, particularly the PST-287K and PST-297S variants. Although the PST-287K variant appears to develop a new 310 helix, the α-helix of PST-297S variant becomes extensively larger, compared with the starting MODELLER predicted structures. This is mainly due to the inclusion of intra-peptide and peptide-solvent interactions in the robust technique of MD simulations. Fig. 7, panels E–G, shows snapshots of representative conformations of the PST peptides during the stable simulation period. The PST-WT structure consisted of a small helix (consisting of Glu-280 to Met-291 residues); the PST-287K structure consisted of a medium helix (consisting of Glu-280 to Val-293); the PST-297S structure consisted of a long helix (consisting of Gly-275 to Val-293). It is, however, important to note that the percentage of conformations displaying the α-helical structures differed among the peptides. Although 66% of the PST-WT conformations contained helical structures, the occurrences of such secondary structures were 40 and 85% for PST-287K and PST-297S, respectively. Interestingly, when the MD simulations were carried out in the presence of TFE, a well known inducer of secondary structures (28), the occurrence of conformations containing the α-helical structures were 85% for PST-WT, 90% for PST-287K, and 85% for the PST-297S peptide. Thus, in general, TFE enhanced the propensity of the PST peptides to assume α-helical structures and the extent of helical structures in these peptides was in the following order: PST-297S > PST-287K > PST-WT.
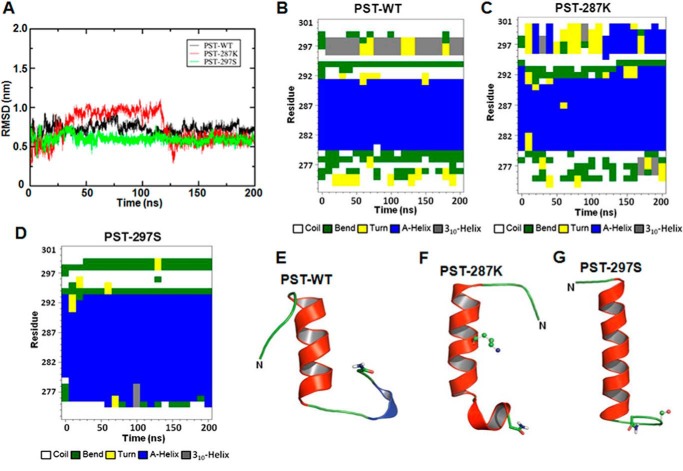
Secondary structure analysis and representative final snapshots of the simulated human PST peptides. Root mean square deviation (RMSD) of backbone atoms of PST peptides as a function of time (panel A). The PST-WT, PST-287K, and PST-297S peptides were homology modeled, well equilibrated, and simulated for 200 ns. The whole trajectory of each peptide was analyzed and changes in the secondary structures were plotted against the simulation time, as shown in the panels: PST-WT (B), PST-287K (C), and PST-297S (D). Final representative conformations of these peptides attained at the end of 200 ns molecular dynamics simulation are shown in the panels: PST-WT (E), PST-287K (F), and PST-297S (G). The peptides are shown in schematic representation, the mutation sites are shown in ball-and-stick format and the amide capping is shown in licorice depiction.
Circular Dichroism Spectroscopic Analysis of Human PST Peptides
To verify the computational predictions regarding structures of the human PST peptides, we carried out CD spectroscopic analysis of these peptides. Spectral deconvolution of CD data suggested the presence of significant amounts of secondary structural elements (α-helix, β-sheet, and β-turn) in all three PST peptides in the physiological buffer (PBS, pH 7.4): PST-WT (27.2%), PST-287K (18.6%), and PST-297S (20.8%) (Fig. 8D); β-sheets contributed the most to the secondary structures. The percentages of β-sheets were different among the peptides, whereas the percentages of β-turns were very similar; percentages of α-helix were also similar among the peptides, each peptide showed a low α-helical content (Fig. 8D).
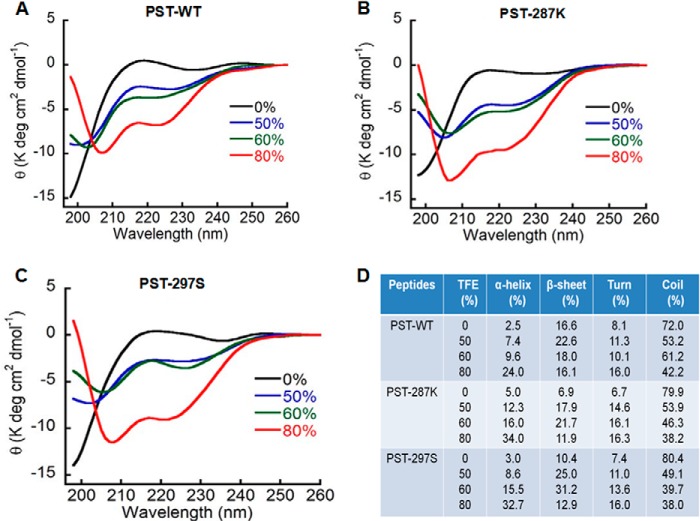
CD spectroscopic analysis of the pancreastatin peptides. CD spectra of PST peptides at a concentration of 50 μm in PBS at pH 7.4 were monitored from 198 to 260 nm wavelength in the absence or presence of 50, 60, and 80% TFE. The α-helix formation was induced by increasing TFE concentration, as indicated by two minima at 208 and 222 nm. The experiment was repeated three times and representative graphs for PST peptides: PST-WT (A), PST-287K (B), and PST-297S (C) were shown. The propensities of each PST peptide to form various structural elements (percentage of α-helix, β-sheet, β-turn, and random coil) with various TFE concentrations (D). The peptides displayed differential propensities for formation of secondary structures (specifically, α-helix) in the following order: PST-287K ≥ PST-297S > PST-WT.
In another set of experiments, CD analysis was carried out with increasing percentages of TFE in PBS to investigate the effect of the hydrophobic environment on the extent of secondary structures. The increasing percentages of TFE induced, in general, a gradual increase in the global secondary structures as reflected by the minima at 208 and 222 nm in the spectra (Fig. 8, panels A–C). TFE specifically induced α-helix formation in the cases of PST-287K and PST-297S peptides, but the effect of TFE was significantly less in the case of the PST-WT peptide. For example, at 80% TFE, the order of α-helix content was: PST-287K (34%) ≈ PST-297S (32.7%) > PST-WT (24%) (Fig. 8D). Likewise, at 60% TFE, the α-helical contents for PST-287K and PST-297S were similarly (16 and 15.5%, respectively) higher than that for the PST-WT peptide (9.6%) (Fig. 8D). These observations largely agree with the computational predictions that PST-287K and PST-297S possess a significantly higher helix forming tendency as compared with PST-WT (Fig. 7) and provide experimental evidence for secondary structures of the PST peptides.
Association of G297S Polymorphism with Biochemical Parameters
Because the PST G297S variant occurred in a fairly large section (~13%) of our study population, we probed for associations of various phenotypic parameters with Gly/Gly and Gly/Ser genotype groups. The genotype groups were of comparable age (~40 years) and did not differ in body mass index, systolic/diastolic/mean arterial blood pressures, or heart rate. Several biochemical parameters (viz. hemoglobin, sodium, potassium, urea, creatinine, triglycerides, HDL, and LDL levels) also did not differ between the genotype groups. However, the Gly/Ser individuals showed significantly higher plasma glucose (by ~17 mg/dl) and higher cholesterol (by 12 mg/dl) than the Gly/Gly individuals (Table 2). Because our study population did not have an equivalent sex representation (~76% males, ~24% females), we also analyzed the phenotypic data for Gly/Ser and Gly/Gly genotype groups after stratification of the subjects by sex. Although the higher glucose effects for the Gly/Ser genotype persisted in male and female groups, these effects did not achieve statistical significance, perhaps because of the decrease in the number of subjects in each group (Fig. 9). Thus, the Ser-297 allele, in general, was associated with two important biochemical traits relevant for metabolic disorders.
TABLE 2
Association of phenotypic parameters with PST-Gly297Ser genotypes.
Data are shown as mean ± S.E. The abbreviations used are: BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; MAP, mean arterial pressure; HDL, high density lipoprotein; LDL, low density lipoprotein. The glucose values are random blood sugar levels. Statistical significance between Gly/Ser and Gly/Gly genotype groups was determined by Levene's test for equality of variances and t test for equality of means using SPSS software (SPSS Inc., Chicago, IL). A p value of <0.05 was chosen for statistical significance between the groups. The significantly differing parameters are shown in bold.
| Parameters | Gly/Gly | Gly/Ser | p value |
|---|---|---|---|
| Demographic/physical | |||
    Age (years) Age (years) | 41.6 ± 0.4 (n = 355) | 39.4 ± 1.1 (n = 55) | |
    Sex (male/female) Sex (male/female) | 65.6%/20.98% | 10.24%/3.17% | |
    BMI (kg/m2) BMI (kg/m2) | 24.5 ± 0.1 (n = 332) | 23.9 ± 0.3 (n = 55) | 0.083 |
| Physiological | |||
    SBP (mm Hg) SBP (mm Hg) | 149.4 ± 1.0 (n = 342) | 146.9 ± 2.9 (n = 55) | 0.436 |
    DBP (mm Hg) DBP (mm Hg) | 90.0 ± 0.6 (n = 340) | 88.0 ± 1.4 (n = 53) | 0.223 |
    MAP (mm Hg) MAP (mm Hg) | 110.2 ± 0.7 (n = 342) | 107.7 ± 1.9 (n = 55) | 0.183 |
    Heart rate (beats/min) Heart rate (beats/min) | 77.9 ± 0.4 (n = 355) | 75.7 ± 1.0 (n = 55) | 0.064 |
| Biochemical | |||
    Hemoglobin (g/dl) Hemoglobin (g/dl) | 13.3 ± 0.1 (n = 353) | 13.7 ± 0.2 (n = 54) | 0.146 |
    Sodium (meq/liter) Sodium (meq/liter) | 137.0 ± 0.3 (n = 354) | 137.6 ± 0.7 (n = 55) | 0.352 |
    Potassium (meq/liter) Potassium (meq/liter) | 4.0 ± 0.02 (n = 342) | 4.0 ± 0.05 (n = 55) | 0.721 |
    Urea (mg/dl) Urea (mg/dl) | 25.6 ± 0.4 (n = 353) | 25.2 ± 1.0 (n = 55) | 0.706 |
    Creatinine (mg/dl) Creatinine (mg/dl) | 0.9 ± 0.01 (n = 346) | 0.9 ± 0.03 (n = 54) | 0.839 |
    Glucose (mg/dl) Glucose (mg/dl) | 104.8 ± 1.5 (n = 327) | 121.9 ± 7.4 (n = 49) | 0.028 |
    Total cholesterol (mg/dl) Total cholesterol (mg/dl) | 158.0 ± 2.0 (n = 349) | 170.0 ± 5.8 (n = 54) | 0.041 |
    Triglycerides (mg/dl) Triglycerides (mg/dl) | 138.0 ± 2.7 (n = 330) | 137.3 ± 6.4 (n = 51) | 0.944 |
    HDL (mg/dl) HDL (mg/dl) | 36.5 ± 0.4 (n = 345) | 37.7 ± 1.0 (n = 54) | 0.266 |
    LDL (mg/dl) LDL (mg/dl) | 82.6 ± 1.4 (n = 334) | 88.7 ± 4.3 (n = 51) | 0.134 |
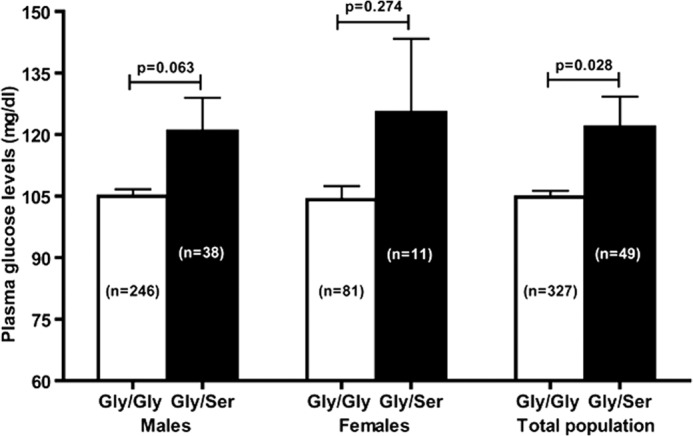
Allele-specific association of the plasma glucose level in vivo. Data are shown as mean ± S.E. Statistical significance between the groups was determined by Levene's test for equality of variances and t test for equality of means using SPSS software (SPSS Inc.). The plasma glucose levels in Gly/Ser individuals (in males as well as females) were higher than Gly/Gly individuals. In the overall study population, Gly/Ser subjects displayed significantly elevated (by ~17 mg/dl) plasma glucose level.
DISCUSSION
Genetic Variations within the PST Peptide
In view of several recent studies implicating important regulatory roles for the CHGA-derived peptide PST in physiology as well as pathophysiology of glucose and insulin homeostasis (1, 9,–11, 15), we set out to probe for functional genetic variations within this peptide in an Indian population. Re-sequencing of 410 subjects led to the discovery of three non-synonymous variants: R253W, E287K, and G297S (Fig. 1). Comparison of amino acid sequences of the PST domain among mammalian species showed that although the minor allele Trp at the 253rd residue position occurred in some non-primates, the Lys residue at 287 or the Ser residue at 297th position was not present in any of the mammals in the course of evolution (Fig. 2). Interestingly, a previous study in a Southern California population detected three genetic variants within the PST domain: R253W, A256G, and G297S (16). Therefore, the present study discovered a novel PST variant (viz. E287K) and did not detect the A256G variant (that was the most frequent variant with minor allele frequency of ~1.1% in the Southern California population). The other PST variants detected in various World populations (Table 1) (E274K (rs371137176), E288K (rs140089514), R300Q (rs147297806)) were also absent in our Indian study population. Interestingly, the Ser-297 allele frequency was almost the same between our study population and Gujarati Indians living in Houston, TX, and this genetic variation appears to be the most frequent within the PST sequence in the overall World population (Table 1). Thus, there are significant differences in the occurrence (presence or absence) as well as allele frequencies of genetic variations in the PST domain among different ethnic/geographically distinct populations.
Differential Activities of the PST Peptides
Among the three discovered variations in the PST peptide, E287K (a change from acidic to basic amino acid) and G297S (a change from the simplest amino acid to one containing aliphatic hydroxyl group) occur within the functional and evolutionarily conserved 29-residue carboxyl terminus (viz. CHGA-(273–301) domain) of the peptide. Therefore, we compared the activities of PST-WT with PST-287K and PST-297S peptides for various cellular processes.
First, we monitored the inhibitory effect of the peptides on insulin-stimulated glucose uptake because PST was reported to diminish glucose uptake in mouse and rat primary adipocytes (8, 11). The rank order of efficacy to diminish insulin-stimulated glucose uptake in differentiated mouse 3T3-L1 adipocytes was: PST-297S (~73%) > PST-287K (~63%) > PST-WT (~23%). This is consistent with a previous report documenting significantly higher potency (~6-fold) and efficacy (~2-fold) of PST-297S over the PST-WT to diminish insulin-stimulated glucose uptake in primary rat adipocytes (10). Thus, although each of the PST peptides blunted glucose uptake, the effect is more pronounced in the case of PST-287K and PST-297S. The exaggerated blockade of glucose uptake by the PST variants might be due to their differential interactions (as compared with the wild-type PST peptide) with the hitherto unknown metabotropic G-protein coupled receptor (29) causing diminished translocation of glucose transporter type-4 (GLUT-4), the major GLUT responsible for insulin-mediated glucose transport (30).
Next, we asked whether the PST peptides differ in their ability to generate NO in the skeletal muscle that is the major site for impaired glucose disposal under insulin resistance conditions (31). Interestingly, the peptides displayed the following order of efficacy to produce NO in L6 cells (NO index values are shown in parentheses): PST-297S (7.21) > PST-287K (5.58) >PST-WT (4.55). As shown in Fig. 4, effects of each of the PST peptides were completely blocked by l-NAME, a potent blocker of NOS, suggesting enzymatic generation of NO by NOS from l-arginine (32, 33) and not by a non-enzymatic method (that may occur during ischemia) from nitrite and nitrate (34). Of note, the PST peptides also exhibited similar effects of NO production in liver HepG2 cells (data not shown), an observation in line with complete restoration of the hepatic NO level (that is ~40% lower in Chga knock-out mice that lacks PST) by PST supplementation in vivo (11). Because the PST effect on NO production is quite prompt (within minutes), it is likely to be via a receptor-mediated increase in intracellular Ca2+ levels, causing NOS activation. Therefore, the differential activities of the PST peptides might be due to altered levels of intracellular Ca2+ triggered by them. Indeed, PST-297S and PST-287K raised the intracellular Ca2+ levels 3.8- and 3.1-fold, respectively, higher than the PST-WT in the skeletal muscle cells (Fig. 5D). This is consistent with the previous report of porcine PST-evoked increase in [Ca2+]i in cultured pancreatic cells (35). Interestingly, the rise in [Ca2+]i by PST peptides in L6 cells was not blocked in the absence of extracellular Ca2+ (Fig. 5, panels A-D), suggesting that PST caused mobilization of Ca2+ from certain intracellular Ca2+ stores. This finding is also consistent with a previous report showing that PST raised the [Ca2+]i by release of Ca2+ from an inositol 1,4,5-triphosphate-sensitive Ca2+ pool (35).
We also investigated the effect of PST peptides on the transcriptional activation of hPepck-1 and hG6Pase, rate-limiting enzymes in the gluconeogenesis pathway, in HepG2 and L6 cells because liver is the site for the highest level of gluconeogenesis in mammals (36) and skeletal muscle is also the site of glyceroneogenesis, a different version of gluconeogenesis (37, 38). As shown in Fig. 6, PST-297S and PST-287K activated the hPepck-1 and hG6Pase promoters to greater extents than the PST-WT. The more pronounced activation of hepatic gluconeogenesis by these PST variants in comparison with the wild-type peptide may be attributed to the stronger effects of these peptides on NO production (Fig. 4) in accordance with previous studies on NO-mediated gluconeogenesis (11). It is noteworthy that PST-WT has been reported to restore Pepck-1 and G6Pase mRNA levels in Chga knock-out mice (11) in corroboration with our promoter activation data (Fig. 6).
Thus, the potencies/efficacies of PST peptides for the different cellular processes, in general, were in the same rank order: PST-297S > PST-287K > PST-WT. Moreover, the magnitudes of effects correlated well across various processes. For example, the correlation coefficient between intracellular Ca2+ level and NO production was 0.9908 with p = 0.0431; likewise, the correlation coefficient between the intracellular Ca2+ level (under Ca2+-free buffer condition) and gluconeogenesis (measured as hPepck-1 transcription) was 0.9998 with p = 0.0056 (Fig. 10). Hence, these cellular processes may be mediated by the same receptor and the crucial amino acid residues within PST (that interact with the receptor) are likely to be similar across these three processes, at least for the PST peptides included in this study.
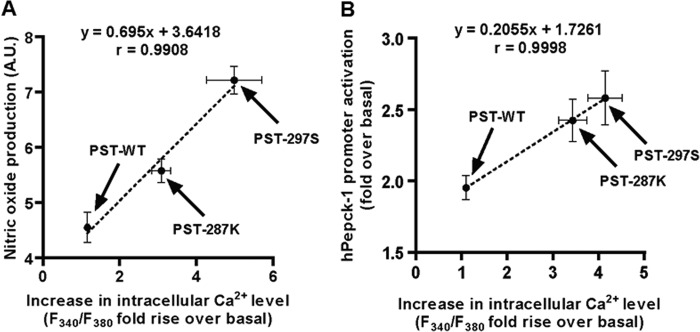
Positive correlations among different cellular processes exhibited by PST peptides. The rise in intracellular Ca2+ levels (in the presence of extracellular Ca2+ in bathing solution) and nitric oxide production showed a strong positive correlation (r = 0.9908, p = 0.0431 with R2 of 0.9817) (panel A); likewise, intracellular Ca2+ levels (in the absence of Ca2+ in bathing solution) and augmentation of hPepck-1 transcription (gluconeogenesis transcription) were significantly correlated (r = 0.9998, p = 0.0056 with R2 of 0.9997) (panel B).
Secondary/Tertiary Structures of the Human PST Peptides
Although the primary structure of PST (initially determined by sequencing of the peptide and then deduced from the cDNA sequence of its parent protein CHGA) in various species has been known for a long time (3, 39, 40) no information on its secondary/tertiary structure is available in the literature. The PST-WT sequence does not have homology with any known peptide family although it shares the Glu-Glu-Glu-Glu-Glu motif (residues 286–290) with gastrin and the carboxyl-terminal Arg-Gly-NH2 with vasopressin (5); our computational analysis showed a similar Glu-Glu-Glu-Glu-Glu motif in mucosal addressin cell adhesion molecule-1 (OMIM code 102670). Therefore, in this study, we attempted to model the structures of PST-WT as well as PST-287K and PST-297S peptides. The structure of PST-WT was modeled using the crystal structure of myocyte-specific enhancer factor 2b as template. The modeled PST-WT consisted of an α helix (spanning Glu-280 to Met-291 residues) and random coils (spanning Pro-273 to Gln-279, Ala-292 to Pro-295 and Phe-299 to Gly-301 residues); the extent of helicity is higher in PST-287K and PST-297S peptides as compared with the PST-WT (Fig. 7). This computational prediction corroborates well with CD data (Fig. 8), especially in the presence of TFE that is known to promote helix formation and provide membrane-mimicking environment (28). Are the structural features attained in the presence of TFE physiologically relevant? Strong correlation between TFE-induced structures and protein structures has previously been established (28). Small peptides with an intrinsic ability to attain helical conformation may adopt very transient/unstable partial helix or random-coil structure in water due to the solvation of peptide backbone. Many peptides also undergo secondary structural transition in the vicinity of the cellular membrane only. CD data in the presence of TFE clearly showed that the PST peptides differ in their propensity to adopt a α-helical structure (Fig. 8), indicating that these peptides are not able to form a stable helix in completely aqueous medium and need a helix-favoring environment. It is also possible that these peptides adopt a helical conformation very slowly in the aqueous environment and require an additive to favor the kinetics. Thus, MD simulations coupled with CD spectroscopy suggested that the PST peptides differ in their intrinsic ability to adopt a α-helical structure, especially, in a hydrophobic/membrane-mimicking environment that the peptides are likely to encounter at lipid bilayers of the cell membranes.
Of note, besides the 29-amino acid form (CHGA-(273–301)) human PST also occurs in several other forms including a 48-amino acid peptide (CHGA-(254–301)) and a 52-amino acid peptide (CHGA-(250–301)), all containing the evolutionarily conserved active C-terminal part of the molecule (41,–43). Among these different molecular forms of PST, the 52-residue peptide has been shown to be the major peptide in human plasma (44). Therefore, we asked whether the secondary structural features that occur in the 29-residue PST peptides are retained in the full-length 52-residue PST. Interestingly, the 52-residue forms of PST-WT, PST-287K, and PST-297S contained similar α helical motifs (data not shown) as noticed in the cases of the 29-residue forms; this observation is consistent with the very much similar activities (at least for inhibition of glucose-stimulated insulin release from isolated rat pancreas) of the carboxyl-terminal fragment and the full-length peptide (3). The structural elements in the carboxyl terminus of the peptide may, hence, be important for its interaction with the hitherto unknown PST receptor.
Correlation between the Human PST Peptide Structures and Their Activities
As shown in Figs. 33–6, the PST variant peptides (viz. PST-287K and PST-297S) exhibited higher potency and efficacy for various cellular processes as compared with the wild-type peptide (PST-WT). Inspection of the computationally predicted structures of these peptides (Fig. 7) along with CD spectroscopy under hydrophobic/membrane-mimicking conditions (Fig. 8) suggested that the variants may contain a greater extent of secondary structure elements (especially, α-helix). Thus, the gain in potency of the variant peptides appears to be associated with their higher α helical contents that might cause altered interactions with the PST receptor.
It may be noted that calculation of the Gribskov score, which predicts if a mutation could be deleterious or neutral, based on a position-specific weighted profile from multiple sequence alignment data (45), showed large differences in the scores between PST-WT and PST variants (E287K,+9.23 to +2.59; G297S, +13.00 to +5.59), indicating that both mutations are deleterious. Further analysis of these PST variants in a recently developed web-server, HANSA, which classifies the deleterious and neutral mutations by position-specific information, local protein structure information, and amino acid properties (46), also predicted these amino acid substitutions as unfavorable. Thus, computational analyses coupled with experimental data of PST secondary/tertiary structures and functions showed a good correlation between the structures and functions of these peptides.
Conclusions and Perspectives
Systematic polymorphism discovery in an Indian population led to the detection of a common variant (PST-297Ser) and a rare novel variant (PST-287Lys) within the functionally active domain of PST. Both variants showed more enhanced activity than the PST-WT in diminishing insulin-stimulated glucose uptake, in increasing intracellular NO and Ca2+ levels, as well as in augmenting transcription of key enzymes for gluconeogenesis. The enhanced activity of the variants for several anti-insulin functions correlated well with their enhanced propensity to adopt a more ordered secondary structures than the wild-type peptide in a membrane-mimicking environment, providing a structural basis for their differential potencies/efficacies. Because increased gluconeogenesis contributes to the pathogenesis of diabetes (47,–50) and the common PST variant PST-297Ser causes significantly higher activation of gluconeogenic enzymes (especially, hG6Pase), carriers of this peptide (viz. ~13% people of Indian origin and perhaps ~300 million people worldwide) may be at the higher risk of developing diabetes. Indeed, human subjects carrying the PST-297Ser allele showed significantly higher plasma glucose level than those with the wild-type PST allele (Fig. 9). Therefore, this study has quantitative implications for inter-individual variations in glucose homeostasis and hence in the pathophysiology of metabolic disorders.
Acknowledgments
We acknowledge Drs. Madhulika Dixit, D. Karunagaran, and Rama Shanker Verma for providing access to some of their equipment. We also thank Rushendhiran Kesavan, S. Abel Arul-Nathan, Niranjana Prabhakar B, Vincent Gerard Francis, Prasanna Vidyasekar, Kalyani Ananthamohan, Lakshmi Subramanian, Bhavani Shankar Sahu, Kiranmayi Malapaka, and Abrar Ali Khan from the Department of Biotechnology, IIT Madras, for their help in this study. We also thank the high performance computational facility at IIT Madras.
*This work was supported in part by the Department of Biotechnology and Science and Engineering Research Board, Government of India (to N. R. M.).
3The abbreviations used are:
- PST
- pancreastatin
- CHGA
- chromogranin A
- PST-297S
- Ser-297 peptide
- PST-WT
- wild-type peptide
- PST-287K
- Lys-287 variant
- hPepck-1
- human phosphoenol pyruvate carboxykinase 1
- pGLuc-Basic vector
- Gaussia luciferase reporter vector
- hG6Pase
- human glucose-6-phosphatase
- TFE
- trifluoroethanol
- l-NAME
- NG-nitro-l-arginine methyl ester
- MD
- molecular dynamics
- CD
- circular dichroism
- ANOVA
- analysis of variance.
REFERENCES
Articles from The Journal of Biological Chemistry are provided here courtesy of American Society for Biochemistry and Molecular Biology
Full text links
Read article at publisher's site: https://doi.org/10.1074/jbc.m113.520916
Read article for free, from open access legal sources, via Unpaywall:
http://www.jbc.org/content/289/7/4455.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1074/jbc.m113.520916
Article citations
NADPH Oxidase 3: Beyond the Inner Ear.
Antioxidants (Basel), 13(2):219, 08 Feb 2024
Cited by: 0 articles | PMID: 38397817 | PMCID: PMC10886416
Review Free full text in Europe PMC
Chromogranin A and its derived peptides: potential regulators of cholesterol homeostasis.
Cell Mol Life Sci, 80(9):271, 29 Aug 2023
Cited by: 0 articles | PMID: 37642733 | PMCID: PMC11072126
Review Free full text in Europe PMC
Chromogranin A-derived peptides pancreastatin and catestatin: emerging therapeutic target for diabetes.
Amino Acids, 55(5):549-561, 13 Mar 2023
Cited by: 4 articles | PMID: 36914766
Review
The immunomodulatory functions of chromogranin A-derived peptide pancreastatin.
Peptides, 158:170893, 13 Oct 2022
Cited by: 1 article | PMID: 36244579 | PMCID: PMC10760928
Review Free full text in Europe PMC
Chromogranin A and its fragments in cardiovascular, immunometabolic, and cancer regulation.
Ann N Y Acad Sci, 1455(1):34-58, 06 Oct 2019
Cited by: 42 articles | PMID: 31588572 | PMCID: PMC6899468
Review Free full text in Europe PMC
Go to all (10) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Diseases
- (1 citation) OMIM - 102670
Nucleotide Sequences (3)
- (1 citation) ENA - BAE01874
- (1 citation) ENA - AEB41037
- (1 citation) ENA - ELR47684
Protein structures in PDBe
-
(1 citation)
PDBe - 1TQEView structure
RefSeq - NCBI Reference Sequence Database (2)
- (1 citation) RefSeq - NM_002591
- (1 citation) RefSeq - NM_001275
SNPs (Showing 7 of 7)
- (2 citations) dbSNP - rs140089514
- (2 citations) dbSNP - rs147297806
- (2 citations) dbSNP - rs371137176
- (1 citation) dbSNP - ss748775092
- (1 citation) dbSNP - rs9658664
- (1 citation) dbSNP - rs9658663
- (1 citation) dbSNP - rs9658662
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Functional Gly297Ser Variant of the Physiological Dysglycemic Peptide Pancreastatin Is a Novel Risk Factor for Cardiometabolic Disorders.
Diabetes, 71(3):538-553, 01 Mar 2022
Cited by: 4 articles | PMID: 34862200
Pancreastatin is an endogenous peptide that regulates glucose homeostasis.
Physiol Genomics, 45(22):1060-1071, 24 Sep 2013
Cited by: 12 articles | PMID: 24064537
Review
Pancreastatin: multiple actions on human intermediary metabolism in vivo, variation in disease, and naturally occurring functional genetic polymorphism.
J Clin Endocrinol Metab, 90(9):5414-5425, 14 Jun 2005
Cited by: 52 articles | PMID: 15956083
Discovery of a novel target for the dysglycemic chromogranin A fragment pancreastatin: interaction with the chaperone GRP78 to influence metabolism.
PLoS One, 9(1):e84132, 20 Jan 2014
Cited by: 12 articles | PMID: 24465394 | PMCID: PMC3896336





