Abstract
Free full text

Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms
Abstract
Bacterial biofilms present a significant medical challenge because they are recalcitrant to current therapeutic regimes. A key component of biofilm formation in the opportunistic human pathogen Pseudomonas aeruginosa is the biosynthesis of the exopolysaccharides Pel and Psl, which are involved in the formation and maintenance of the structural biofilm scaffold and protection against antimicrobials and host defenses. Given that the glycoside hydrolases PelAh and PslGh encoded in the pel and psl biosynthetic operons, respectively, are utilized for in vivo exopolysaccharide processing, we reasoned that these would provide specificity to target P. aeruginosa biofilms. Evaluating these enzymes as potential therapeutics, we demonstrate that these glycoside hydrolases selectively target and degrade the exopolysaccharide component of the biofilm matrix. PelAh and PslGh inhibit biofilm formation over a 24-hour period with a half maximal effective concentration (EC50) of 69.3 ± 1.2 and 4.1 ± 1.1 nM, respectively, and are capable of disrupting preexisting biofilms in 1 hour with EC50 of 35.7 ± 1.1 and 12.9 ± 1.1 nM, respectively. This treatment was effective against clinical and environmental P. aeruginosa isolates and reduced biofilm biomass by 58 to 94%. These noncytotoxic enzymes potentiated antibiotics because the addition of either enzyme to a sublethal concentration of colistin reduced viable bacterial counts by 2.5 orders of magnitude when used either prophylactically or on established 24-hour biofilms. In addition, PelAh was able to increase neutrophil killing by ~50%. This work illustrates the feasibility and benefits of using bacterial exopolysaccharide biosynthetic glycoside hydrolases to develop novel antibiofilm therapeutics.
INTRODUCTION
Bacterial biofilms provide a protective life-style for bacteria and are extremely challenging and costly to treat because they are notoriously recalcitrant to antibiotics and host defenses (1–5). It is estimated that 65 to 80% of all human bacterial infections are related to biofilms (6). Biofilms are complex communities of bacteria embedded in an extracellular matrix composed of proteins, extracellular DNA (eDNA), and exopolysaccharides. The exopolysaccharide component of the biofilm matrix can function to impair antibiotic penetration (7, 8) and provide a barrier against phagocytosis by host immune cells (9). Given the rise of antibiotic resistance and the discovery that subinhibitory concentrations of antibiotics and antimicrobial compounds can promote biofilm formation, there is an urgent need for novel and effective treatments that target and disrupt biofilms (10–12).
Pseudomonas aeruginosa is a ubiquitous, Gram-negative, opportunistic pathogen that is commonly associated with nosocomial infections (13). Mortality associated with P. aeruginosa infections is high (14), and the emergence of multidrug resistance and even pandrug resistance to antimicrobials has been reported (15). The ability of P. aeruginosa to form biofilms is thought to be an important factor underlying the success of this organism in causing persistent infections in humans. Because the biofilm matrix is critical to the persistence of and resistance to antimicrobial agents (7, 8, 16, 17), studies have focused on developing prophylactic treatments that inhibit biofilm formation through the activation of intrinsic bacterial responses (18–23). However, most compounds are unable to disrupt established biofilms, which is a more clinically relevant condition. Only nitric oxide (24), cis-2-decenoic acid (25), and antibiofilm peptide 1080 (26) have been demonstrated to mediate both P. aeruginosa biofilm prevention and disruption. These molecules have only been tested against the P. aeruginosa PAO1 strain and require extended incubation times (≥24 hours) to be efficacious against established biofilms, and their lack of specificity may exert negative effects on the natural microbiota.
An alternate approach to the treatment of established biofilms is the use of therapeutic enzymes that degrade the biofilm matrix. Dornase alfa (deoxyribonuclease I) is the only enzyme in clinical use that disrupts P. aeruginosa biofilms. This therapeutic enzyme functions by hydrolyzing the eDNA within the extracellular matrix (27, 28). Because eDNA is involved in initial biofilm establishment, immature P. aeruginosa biofilms are more sensitive to deoxyribonuclease I treatment than are mature biofilms (29, 30). This decrease in the deoxyribonuclease I sensitivity of mature biofilms is thought to reflect the increased production and utilization of exopolysaccharides in these biofilms. The glycoside hydrolase DspB (DispersinB), which hydrolyzes the biofilm exopolysaccharide poly-β-1,6-N-acetyl-d-glucosamine (PNAG/PIA), has also shown promise as a therapeutic enzyme (31–33). PNAG is required for biofilm formation and integrity of several Gram-positive and Gram-negative pathogenic bacteria but is not present in P. aeruginosa biofilms (34–39). P. aeruginosa has the genetic capacity to synthesize at least three different biofilm exopolysaccharides: Psl, Pel, and alginate. These polysaccharides are integral components of the extracellular biofilm matrix (40, 41). Although alginate production results in a mucoid phenotype and is correlated with chronic infection and poor prognosis in patients with cystic fibrosis, this exopolysaccharide is dispensable for biofilm formation in nonmucoid P. aeruginosa strains (42–44). Psl is a neutral polysaccharide composed of a pentasaccharide repeat unit of d-mannose, l-rhamnose, and d-glucose (45), whereas Pel has been recently identified as a cationic polysaccharide composed of partially deacetylated N-acetyl-d-glucosamine and N-acetyl-d-galactosamine (46, 47). Psl and, under some circumstances, Pel function to facilitate initial surface attachment (45, 46, 48–50). Both exopolysaccharides play a significant role in the formation and maintenance of the biofilm architecture (7, 51, 52). P. aeruginosa strains with genetic deletions of the pel and psl operons are profoundly impaired in biofilm formation and virulence in a mouse model of acute infection (43). Psl provides protection against neutrophil phagocytosis and antibiotics with diverse biochemical properties (8, 9), whereas Pel enhances resistance to aminoglycosides (7, 16). Although the preference of Pel or Psl is often strain-specific, many isolates are capable of switching between the synthesis of Pel and that of Psl in response to stress to maintain infection in the host (53, 54). This adaptive mechanism underscores the importance of developing therapies that target both exopolysaccharides.
We therefore sought to identify enzymes that selectively target and degrade Psl and Pel. One common feature shared by many exopolysaccharide biosynthetic operons is the presence of a gene encoding a glycoside hydrolase that is proposed to hydrolyze the exopolysaccharide produced by the biosynthetic pathway (47, 55–59). We have exploited these naturally derived glycoside hydrolases as a method of biofilm prevention and disruption. We demonstrate that the addition of low nanomolar concentrations of the enzymes PelAh and PslGh can prevent biofilm formation and disrupt existing biofilms of laboratory, clinical, and environmental isolates of P. aeruginosa in vitro at nanomolar concentrations. In addition to disrupting biofilms, these noncytotoxic enzymes potentiate antibiotics and enhance susceptibility to killing by neutrophils. These studies provide us with a method to find enzymes with antibiofilm activity for the treatment and eradication of chronic bacterial infections.
RESULTS
The isolated glycoside hydrolase domains of PelA and PslG are soluble enzymes
Previous bioinformatics analyses have identified PslG and the N-terminal domain of PelA as putative periplasmic glycoside hydrolases encoded in the psl and pel biosynthetic operons, respectively (47). We recently purified and functionally characterized PslG31–442, a member of glycoside hydrolase family 39, herein referred to as PslGh (60). This construct removes an N-terminal transmembrane domain, producing a soluble, catalytically active glycoside hydrolase domain that can hydrolyze Psl. PelA is a multifunctional protein that contains at least two catalytic domains—a putative glycoside hydrolase domain and a CE4 deacetylase domain (47). On the basis of bioinformatics prediction using the CAZymes Analysis Toolkit (61), we generated a PelA47–303 construct, herein referred to as PelAh, to explicitly study the activity of the glycoside hydrolase domain. This construct was soluble and could be purified to homogeneity with nitrilotriacetic acid (Ni-NTA) purification and size exclusion chromatography, which yield 50 mg of protein per liter of bacterial cell culture.
Glycoside hydrolases catalyze the disruption of biofilms
We hypothesized that the exogenous application of the glycoside hydrolases PelAh and PslGh to Pel- and Psl-dependent biofilms, respectively, would result in hydrolysis of the exopolysaccharides, thereby disrupting the established biofilms. To assay for biofilm disruption, we produced biofilms using the following strains: PA14 (Pel-dependent matrix); PAO1 (Psl-dependent matrix); and l-arabinose–inducible P. aeruginosa PAO1 ΔwspF Δpsl PBADpel and PAO1 ΔpelF PBADpsl, which exclusively produce Pel and Psl, respectively. PelAh and a putative catalytically inactive E218A variant (PelAh E218A) were applied to Pel-dependent biofilms, whereas PslGh and an inactive E165Q/E276Q variant (PslGh E165Q/E276Q) were applied to Psl-dependent biofilms. Images captured with confocal microscopy coupled with fluorescence-labeled lectins from Hippeastrum hybrid Amaryllis (HHA; specific for Psl) and Wisteria floribunda (WFL; specific for Pel) demonstrated that catalytically active hydrolases, but not the inactive variants, were capable of degrading the Pel- and Psl-dependent biofilm biomass on the basis of the elimination of fluorescence signal following treatment (Fig. 1).
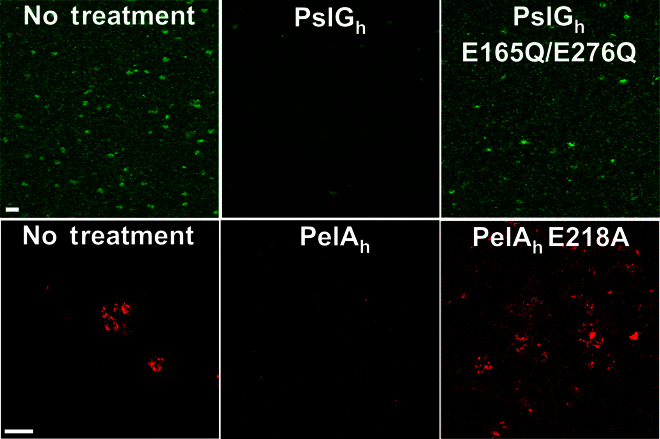
Representative confocal images of Psl biofilms grown statically for 24 hours (top) and Pel biofilms cultivated for 48 hours (bottom) under flow conditions and treated with wild-type hydrolases or hydrolases that have point mutations to catalytic residues. Biofilms were stained with the HHA Psl-specific lectin (green) and WFL Pel-specific lectin (red). Scale bars, 30 μm.
Crystal violet staining was subsequently utilized to quantify the effect of hydrolase treatment on the total biofilm biomass. A 2-hour treatment of a Pel-dependent biofilm with PelAh resulted in disruption of 99% of the biomass, whereas both the PelAh E218A variant and PslGh, added in 100-fold excess relative to PelAh, exhibited no significant difference compared to that of the untreated biofilm (Fig. 2A). Similar results were obtained for Psl-dependent biofilms wherein only the treatment with a catalytically active PslGh resulted in a 98.5% reduction in biofilm biomass. The activity of both enzymes was observed to be dose-dependent. When incubated with established biofilms for 1 hour, PelAh exhibited a half maximal effective concentration (EC50) of 35.7 ± 1.1 nM, whereas PslGh had an EC50 of 12.9 ± 1.1 nM (Fig. 2B). Time course experiments using fixed concentrations of PelAh and PslGh revealed a continuous decrease in biofilm biomass over time (fig. S1). The activity of both enzymes was minimally affected by the presence of serum with observed EC50 of 66.1 ± 1.2 nM for PelAh and 6.7 ± 1.1 nM for PslGh (fig. S2). Combined, these results indicate that biofilm disruption is catalytic, rapid, and exopolysaccharide-specific.
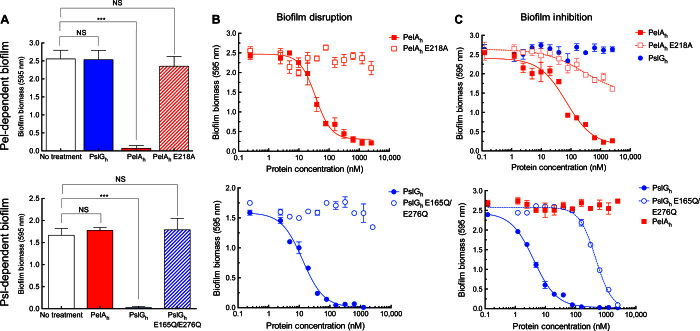
(A) Crystal violet staining of biofilms following the exogenous addition of glycoside hydrolases or catalytic variants. (B) Dose-response curves to examine the disruption of biofilm biomass by the exogenous treatment of each glycoside hydrolase and variant. (C) Dose-response curves to examine the prevention of biofilm biomass in the presence of various glycoside hydrolases. Each data point represents the mean from three independent experiments of n = 3 crystal violet microtiter plate wells. EC50 values were calculated using nonlinear least-squares fitting to a dose-response model. Error bars indicate SEM. ***P ≤ 0.001. NS, no significant difference.
Glycoside hydrolases inhibit biofilm formation but not bacterial growth
Because the glycoside hydrolases were effective at disrupting established biofilms, we next sought to determine whether the application of enzyme to bacterial culture could be utilized as a prophylactic strategy to prevent biofilm formation. The addition of PelAh, but not PslGh, to Pel-producing P. aeruginosa abrogated biofilm formation. Dose titration indicated that Pel biofilms could be prevented over 24 hours by the addition of PelAh with an EC50 of 69.3 ± 1.2 nM (Fig. 2C). As visualized in borosilicate tubes, PelAh prevented pellicle biofilm at the air-liquid interface, and bacterial cells grew exclusively in the planktonic state (fig. S3). Addition of PelAh E218A, which cannot catalyze the disruption of biofilms, resulted in a statistically significant reduction in biofilm biomass at concentrations of ≥500 nM. Although an accurate EC50 value could not be readily determined for this catalytic variant, 5 μM of the enzyme variant (>70 times greater than the EC50 of the wild type) resulted in <50% reduction of the biomass relative to untreated cells. The presence of ≥10 μM PslGh did not affect the ability of P. aeruginosa to form Pel-dependent biofilms. Consistent with the results for PelAh on Pel-dependent biofilms, the addition of 1 μM PslGh to Psl-producing cultures under biofilm-forming conditions resulted in a complete inhibition of biofilm formation, and dose titration indicated that PslGh had an EC50 of 4.1 ± 1.1 nM over 24 hours. To examine whether the effect was the direct result of PslGh activity, we tested the catalytically inactive variant PslGh E165Q/E276Q. This variant was >100-fold less effective in biofilm prevention (EC50 of 466.5 ± 1.1 nM) relative to the catalytically active enzyme. Addition of ≥10 μM PelAh had no effect on Psl-dependent biofilm production.
We next examined the length of time in which the glycoside hydrolases could prevent biofilm formation. A single dose of PelAh or PslGh prevented biofilm formation for 48 and 72 hours, respectively. The formation of a Pel-dependent biofilm at 72 hours was associated with proteolytic degradation of PelAh, whereas no degradation of PslGh was observed over the entirety of the experiment (fig. S4). The growth rate of P. aeruginosa PAO1 exposed to ≥20 μM of either glycoside hydrolase was unaffected over 6 hours of static growth when compared to a no-treatment control (fig. S5). The absence of enzyme cross-reactivity between exopolysaccharides indicates that biofilm inhibition is highly specific. This result, combined with bacterial growth curves, demonstrates that exogenous glycoside hydrolases do not impede biofilm formation by altering cell viability and growth.
Biofilm-disrupting enzymes are noncytotoxic
Because exogenous PelAh and PslGh did not affect P. aeruginosa cell viability and growth, we next sought to examine whether enzyme treatment affected mammalian cells. IMR-90 human lung fibroblast cells treated for 5 hours with concentrations of up to 1 mg/ml of either enzyme, which are ~100-fold above the concentration required for effective biofilm disruption, resulted in no significant difference in cell area or length-to-width ratio (Fig. 3A). Following a 48-hour incubation, no significant difference in cellular viability was observed in PelAh- and PslGh-treated cells regardless of the media used (Fig. 3B). The Clostridium difficile toxin TcdB (62) was used as a control for cell morphology because it results in cell rounding, whereas digitonin, which permeabilizes the cells, was used to monitor cellular viability (Fig. 3B). Western blot analysis of the media confirmed that PelAh and PslGh remained intact for the 48-hour duration of the experiment (fig. S6). Together, these results suggest that the enzymes do not interfere with mammalian cell morphology and viability.
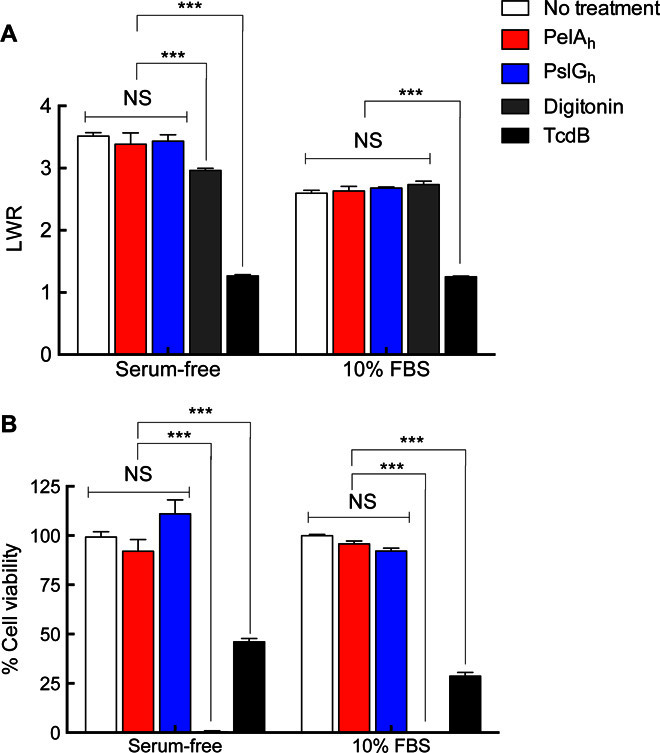
(A) IMR-90 cellomics assay to measure the length-to-width ratio (LWR) of the cells using CellTracker Orange CMRA. (B) IMR-90 fibroblast cell viability assay using PrestoBlue reagent. All data were normalized to a no-treatment control (100%). The C. difficile toxin TcdB was used as a positive control in cell morphology assays, and the detergent digitonin was utilized as a negative control in cell viability assays. Each data point represents the mean from three independent experiments of n = 3 cellomic and PrestoBlue measurements in microtiter plate wells. Error bars indicate SEM. ***P ≤ 0.001. NS, no significant difference.
Enzymes potentiate antibiotics and ameliorate human neutrophil killing
Previous studies have demonstrated that both Pel and Psl enhance antibiotic resistance (7, 8). We therefore theorized that glycoside hydrolase degradation of these polymers could potentiate the activity of antimicrobial agents. Because the antibiotic colistin targets the cell membrane of P. aeruginosa, it is active against both metabolically active and dormant cells found within biofilms (63). The effect of combining glycoside hydrolase treatment with subinhibitory concentrations of colistin was therefore examined. As predicted, the treatment of P. aeruginosa Pel- and Psl-dependent 24-hour biofilm cultures with colistin (50 μg/ml) or those grown in the presence of PelAh or PslGh (2 μM) alone had no effect on the viability of the bacteria (Fig. 4A). However, prophylactic treatment with PelAh or PslGh before treatment with colistin resulted in an approximately 2.5-log reduction in bacterial colony-forming units (CFUs). A 5-hour cotreatment of 24-hour biofilms with either PelAh or PslGh and colistin resulted in a similar reduction in CFUs (Fig. 4B). These results indicate that these glycoside hydrolases are compatible with antibiotics and can potentiate the antimicrobial activity of colistin.
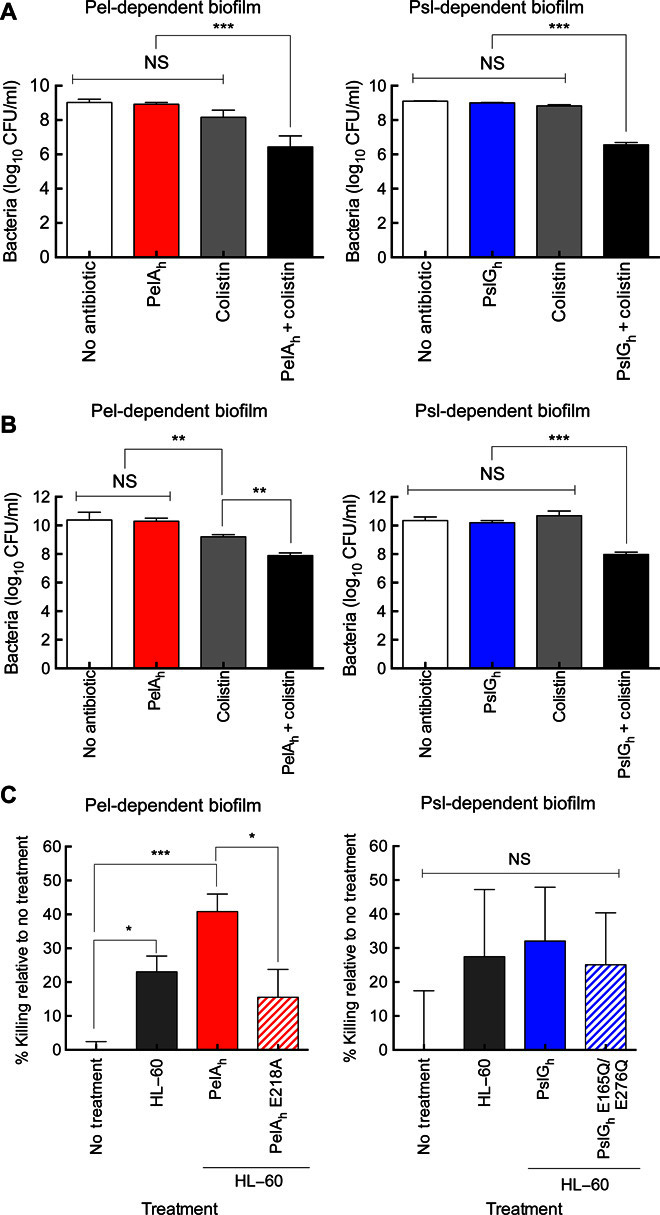
(A) CFUs for PAO1 ΔwspF Δpsl PBADpel (left) and PAO1 ΔpelF PBADpsl (right) following growth in the presence or absence of glycoside hydrolases before treatment with colistin. The mean was calculated from LB agar plate counts from three independent experiments. (B) CFUs for biofilm cultures in (A) after treatment with glycoside hydrolases and colistin for 5 hours. The mean was calculated from LB agar plate counts from three independent experiments. (C) HL-60 neutrophil killing of strain PAO1 ΔwspF Δpsl PBADpel and PAO1 ΔpelF PBADpsl following biofilm formation and treatment with PelAh and PslGh and their catalytic variants, respectively. Percent killing was normalized to a no-treatment control. Error bars indicate SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. NS, no significant difference.
Because P. aeruginosa exopolysaccharides enhance resistance to human neutrophil killing, we next investigated whether glycoside hydrolase treatment could enhance susceptibility to immunomediated killing. To determine whether hydrolase treatment could affect neutrophil killing, we examined the ability of PelAh and PslGh to enhance the susceptibility of P. aeruginosa to the human HL-60–derived neutrophils. Treatment of Pel-dependent P. aeruginosa biofilms with PelAh increased the degree of HL-60–mediated microbial killing from approximately 22 to 42% (Fig. 4C). This was not observed in a PelAh E218A variant, indicating that the enhanced susceptibility to neutrophils is due to the catalytic activity of the enzyme that disrupts the biofilm. This effect was specific to PelAh because neither PslGh nor the E165Q/E276Q variant was significant to ameliorate neutrophil killing or affect neutrophil activity (Fig. 4C). Combined, these data provide further evidence that PelAh and PslGh do not affect mammalian cell function, and that PelAh can function to enhance neutrophil killing of P. aeruginosa.
Enzymes effectively disrupt biofilms from clinical and environmental isolates
Our previous work established that clinical strains of P. aeruginosa can be divided into four different classes on the basis of their dependence on Pel and Psl exopolysaccharides for biofilm formation (53). The effect of PelAh and PslGh on biofilm disruption of isolates from each of the four classes was evaluated. Under the growth and assay conditions tested, treatment with 300 nM PslGh + PelAh for 2 hours resulted in a 70 to 94% disruption of biofilms formed by isolates from classes II to IV (Fig. 5). PslGh was more effective at disrupting biofilms from classes II to IV, as anticipated, because the Psl polysaccharide is a major contributor to the biofilm biomass of these strain classes. An additive effect was observed for the matrix overproducer CF127, wherein the combination of both enzymes led to the largest decrease in biofilm biomass (95% biomass reduction for the combined PslGh + PelAh compared to 85% for PslGh alone). Strains PA14 (the sole member of class I) and CF127 (a class IV member) required 1 μM of enzyme to reduce the biofilm biomass by 58 and 95%, respectively. These data demonstrate that the enzymes are compatible with one another and can be utilized to disrupt biofilms of clinical and environmental P. aeruginosa isolates.
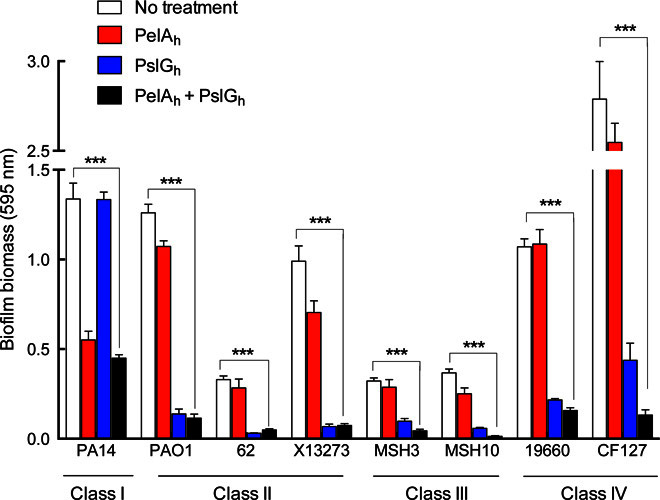
Isolates were grouped into categories as previously described, and the glycoside hydrolases PelAh and PslGh were exogenously added either individually or together and allowed to incubate for 2 hours. All strains were treated with 300 nM of each enzyme, with the exception of PA14 and CF127, which were treated with 1000 nM. Each data point represents the mean from three independent experiments of n = 3 crystal violet microtiter plate wells. Error bars indicate SEM. ***P ≤ 0.001.
DISCUSSION
It is well established that bacterial infections involving biofilm formation are difficult to eradicate because of their enhanced resistance to antimicrobials and host immune defenses. Here, we demonstrate that low nanomolar concentrations of the glycoside hydrolases PslGh and PelAh can prevent and rapidly disrupt biofilms produced by P. aeruginosa in vitro. Developing enzymes that target exopolysaccharides for therapeutic purposes has several advantages, including the key roles that polysaccharides play during matrix development, the short time required for biofilm disruption, the selectivity of the enzymes, and a low potential for the development of acquired resistance.
The exopolysaccharides Pel and Psl bind eDNA and proteins in the matrix to form a cohesive and structurally robust biofilm (46, 64, 65). These interactions do not appear to hinder PelAh and PslGh from accessing and hydrolyzing Pel and Psl, respectively. The production of these extracellular matrix glycans provides protection not only to the cells that synthesize these molecules but also to other bacteria within the biofilm matrix, such as exopolysaccharide nonproducers, metabolically dormant persister cells, and other Gram-positive and Gram-negative pathogens growing in mixed-species biofilms. Pel can promote mixed-species interaction with Staphylococcus aureus (66), and Psl confers protection against detergent stress and antibiotics upon Escherichia coli and S. aureus (8, 67). Because many infections are polymicrobial, targeting and hydrolyzing Pel and Psl may be sufficient to disrupt these bacterial communities and extend treatment to other bacterial species that do not produce these polysaccharides themselves.
Pel and Psl play important roles at multiple stages of P. aeruginosa biofilm development and maturation (53, 68). Therefore, biofilm disruption with PelAh and PslGh is not sensitive to the maturation state of the biofilm, as is the case with deoxyribonuclease I. Penetration and disruption of the biofilm have several potential consequences, including increased penetration of antibiotics within the biofilm matrix and reduction of microenvironments that can render antimicrobials inactive. Our results indicate that PslGh and PelAh are compatible with one another and with antibiotics and neutrophils. Because the enzymes function as adjuvants for the innate immune system and antibiotics, they do not alter growth or metabolism, and therefore place no direct selective pressure on the bacteria. In addition, targeting of extracellular components avoids the myriad of cellular resistance mechanisms used by P. aeruginosa against antimicrobials such as efflux or intracellular drug modification. Neither enzyme was susceptible to proteolytic degradation by the bacteria or mammalian cells for >24 hours and retained catalytic activity. Furthermore, the rapid action of these enzymes avoids the need for prolonged exposure of bacteria to these molecules and should reduce the risk of the emergence of resistance. The specific hydrolysis of Pel and Psl should minimize off-target effects on the microbiota and host carbohydrates on the basis of the unique chemical structures of these exopolysaccharides.
Preclinical studies in animal models are essential to the development of these enzymes as therapeutic agents for the treatment of Pseudomonas infections. Although several mammalian-derived glycoside hydrolases are in clinical use (69), examining immunotolerability to bacteria-derived enzymes in vivo will serve as the next important step. Our in vitro work has demonstrated that both enzymes are noncytotoxic, do not alter the morphology and attachment of mammalian cells, and do not perturb the cellular function of neutrophils, which are the first line of defense of the innate immune system. Challenges with the tolerability of these enzymes could, if required, be mitigated using methodologies such as PEGylation or encapsulation in hydrogels, which have successfully reduced the immunogenicity and antigenicity of other therapeutic enzymes (70, 71).
Because many Gram-positive and Gram-negative bacterial exopolysaccharide biosynthetic operons encode a putative glycoside hydrolase or lyase, it is likely that the strategy we have used could be extended to prevent and disrupt other exopolysaccharide-dependent biofilms. Examples include BcsZ, WssD, PssZ, and PgaB involved in cellulose, acetylated cellulose, Listeria monocytogenes exopolysaccharide, and PNAG biosynthesis, respectively (55, 56, 58, 59). Glycoside hydrolase therapy has the potential to target diverse biofilms from many Gram-positive and Gram-negative bacteria that are extremely relevant to both healthcare and industrial settings. In conclusion, our study demonstrates that components of the P. aeruginosa exopolysaccharide biosynthetic operons can be manipulated to disrupt biofilms produced by the bacterium, allowing for antibiotic potentiation and effective killing by innate immunity.
MATERIALS AND METHODS
Strains
Strains used in this study are reported in table S1, and detailed culture conditions are described below.
Cloning, expression, and purification of PelA and PslG constructs
PslGh was purified as previously described (60). The DNA sequence of pelA from P. aeruginosa PAO1 was obtained from GenBank under accession no. AAG06452.1 (72). The PRED-TAT server (73) indicated that PelA has a TAT (twin-arginine translocation) signal sequence from residues 1 to 45. To obtain soluble protein constructs, we amplified pelA from genomic DNA by polymerase chain reaction using the primers CTGCATATGGGCGGGCCGTCCAGCGTGGCG and TTTCTCGAGTCACGGTTGCAC CTCGACGTC, respectively. Introduced NdeI and XhoI restriction sites were underlined, and each gene was ligated into the pET28a (Novagen) expression vector encoding an N-terminal polyhistidine tag. This generated PelA47–303. Site-directed mutagenesis was performed to generate protein variants, using the QuikChange Lightning Kit according to the prescribed protocol (Agilent Technologies). Generated constructs were verified by sequencing performed by ACGT DNA Technologies Corporation.
E. coli BL21 (DE3) CodonPlus cells (Stratagene) were transformed with the expression plasmid and grown in 2 liters of Lauria-Bertani (LB) broth containing kanamycin (50 μg/ml) at 37°C. When the optical density at 600 nm (OD600) of the cell culture reached 0.5 to 0.6, protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.5 mM. After induction, the cells were incubated overnight at 18°C with shaking at 200 rpm, before being harvested by centrifugation at 5000g for 30 min at 4°C. Cell pellets containing PelA47–303 were resuspended in 40 ml of buffer A [20 mM imidazole, 50 mM tris-HCl (pH 7.5), 300 mM NaCl, and 2% (v/v) glycerol] with one SIGMAFAST Protease Inhibitor Tablet. The cells were lysed by at least three passes through an EmulsiFlex C3 homogenizer at 100 MPa (Avestin Inc.), and the resulting cell debris was separated from soluble protein by centrifugation at 35,000g for 30 min. The supernatant was applied to 5 ml of Ni-NTA Superflow resin packed into a gravity column (Qiagen) preequilibrated with buffer A. The column was washed with 3 CV (column volume) of buffer A, and the expressed protein was eluted with buffer A supplemented with 250 mM imidazole. The eluted fractions were concentrated to a 1- to 2-ml volume using an Amicon Ultra centrifugation filter device (Millipore) with a 10-kDa cutoff, and the protein was further purified by size exclusion chromatography using a HiLoad 16/60 Superdex 200 gel filtration column (GE Healthcare). The protein was judged to be >95% pure by SDS–polyacrylamide gel electrophoresis, and the protein could be concentrated to 8 to 10 mg/ml and stored at 4°C for at least 1 month without precipitation or degradation.
Confocal microscopy
Psl biofilms were grown overnight at room temperature in uncoated 15 μ-Slide VI0.4 flow cell chambers (ibidi GmbH). The channels were inoculated with 200 μl of a culture with an OD600 of 0.5 grown in LB no salt (LBNS) supplemented with 0.5% arabinose. Biofilms were washed three times with sterile phosphate-buffered saline and then treated with PslGh, PslGh E165Q/E276Q, and buffer-only control [50 mM tris (pH 7.5), 150 mM NaCl, and 10% (v/v) glycerol] statically for 1 hour at room temperature. The final enzyme concentration was 86 nM. After digestion, the biofilms were stained with fluorescein isothiocyanate–conjugated HHA lectin (100 μg/ml; EY Laboratories) for 2 hours at 4°C, as previously described by Ma et al. (74). The biofilms were then washed and fixed with 4% paraformaldehyde. Fluorescent images were acquired with an Olympus FV1000 filter confocal system using a 20× LUCPLFLN objective lens with a numerical aperture of 4.5 (Olympus America Inc.). Images were analyzed and constructed using the Olympus FluoView version 03.01 software.
Pel-dependent biofilms were cultivated as previously described (46), with minor modifications. Flow cell chambers were inoculated with a mid-log LB culture of P. aeruginosa PA14 that was diluted with glucose minimal medium (final glucose concentration of 0.3 mM) to an OD600 of 0.01. Cells were allowed to attach for 1 hour before induction of flow. Biofilms were grown on glucose minimal medium for 2 days at room temperature at a constant flow rate (10 ml/hour) before treatment with PelAh, PelAh E218A, and buffer-only control [20 mM Tris (pH 8.0), 150 mM NaCl, and 10% (v/v) glycerol] statically for 1 hour at room temperature. The final enzyme concentration was 85 nM. After digestion, biofilms were washed, and Pel was then stained with fluorescein-labeled WFL lectin (100 μg/ml; Vector Laboratories) for 15 min. Stained biofilms were washed before visualization on a Zeiss LSM 510 confocal laser scanning microscope. Image analysis was conducted using the Velocity software (Improvision). Experiments were performed in biological duplicate.
Microtiter dish biofilm assay
For examination of biofilm prevention, Psl and Pel arabinose–inducible P. aeruginosa PAO1 (PAO1 ΔpelF PBADpsl and PAO1 ΔwspF Δpsl PBADpel) and clinical isolates were grown at 37°C overnight with shaking at 200 rpm. The cultures were normalized to an OD600 of 0.5 and then diluted 1:100 in LBNS. l-Arabinose was added to a final concentration of 0.5% (w/v) to induce exopolysaccharide biosynthesis and biofilm formation. Diluted culture (95 μl) was added to sterile 96-well polystyrene microtiter plates (Thermo Scientific cat. no. 243656), and varying concentrations of PelAh or PslGh (0.1 to 5 μM) were added in 5-μl aliquots to give a final volume of 100 μl. The cultures were incubated statically for 24 hours at 25°C to allow for biofilm formation. For elimination of edge effects, ~200 μl of sterile water was placed in all outside wells, and the plate was sealed with parafilm. After incubation, nonadherent cells and media were removed by thoroughly washing the plate with deionized water. The wells were stained with 150 μl of 0.1% (w/v) crystal violet for 10 min and then rinsed with water. The remaining dye was solubilized by addition of 150 μl of 95% (v/v) ethanol and left for 10 min, after which the absorbance was measured at 595 nm using a SpectraMax M2 spectrophotometer (Molecular Devices). The amount of biofilm was proportional to the absorbance from staining with crystal violet (75).
For biofilm disruption assays, biofilm cultures were grown statically for 24 hours. Following incubation, nonadherent cells and media were removed by washing the plate with distilled water. The wells were filled with 95 μl of 100 mM HEPES sodium buffer (pH 7.0) followed by 5 μl of varying concentrations of each hydrolytic enzyme (2 to 5 μM). Reactions were allowed to proceed for up to 60 min at 25°C on a rotating nutator, at which time the reaction was quenched by washing the plates with distilled water. The wells were stained with 150 μl of 0.1% (w/v) crystal violet for 10 min and then washed and solubilized with ethanol before quantification. All reactions were completed in triplicate. The addition of kanamycin (2.5 mg/ml) to the culture before biofilm formation was used as a positive control.
P. aeruginosa growth assay
To assay for glycoside hydrolase cytotoxicity to P. aeruginosa PAO1, we set up a bacterial growth assay as described for the biofilm inhibition assay, with 25 μM PelAh or PslGh added at the time of inoculation. The bacteria were grown statically at 37°C in a thermocontrolled SpectraMax M2 spectrophotometer. At 30-min intervals, the OD600 of each culture was measured for a duration of 6 hours. An untreated culture was used as a control.
Antibiotic susceptibility assay
Overnight cultures of P. aeruginosa PAO1 ΔwspF Δpsl PBADpel (Pel-dependent) and P. aeruginosa PAO1 ΔpelF PBADpsl (Psl-dependent) were diluted to an OD600 of 0.05 in LBNS with 0.5% (w/v) l-arabinose and were grown statically in polystyrene tubes at a final volume of 1 ml per tube. For prophylactic treatment, 2 μM PelAh or PslGh was added at the time of inoculation and incubated with the bacteria for 24 hours at 25°C. Following incubation, colistin was added at a final concentration of 50 μg/ml and was incubated for 24 hours. For treatment of 24-hour biofilms, 2 μM PelAh or PslGh was added concurrently with a final colistin concentration of 50 μg/ml and was incubated with the biofilm at 25°C for 5 hours. Cultures that were not subjected to treatment or were only treated with enzymes served as controls. Adherent biofilm and embedded cells were resuspended by scraping the tubes and vigorous pipetting to ensure removal of all cellular materials and to prevent experimental bias. Viability was quantified by serial dilutions and CFU counts on LB agar plates of the surviving population. Experiments were performed three times to obtain both mean and SE.
Human neutrophil killing assay
Overnight cultures of P. aeruginosa PAO1 ΔwspF Δpsl PBADpel and P. aeruginosa PAO1 ΔpelF PBADpsl were diluted to an OD600 of 0.05 in LBNS with 0.5% l-arabinose and inoculated in a 96-well tissue culture–treated plate at a final volume of 100 μl per well. The plate was incubated statically at 28°C for 20 hours. Supernatants were aspirated, and 100 μl of phenol red–free RPMI medium + 10% fetal bovine serum (FBS) containing 0.5 μM PelAh or PslGh was added. The plate was incubated at room temperature on the nutator for 1 hour. Following pretreatment with hydrolase, 100 μl of RPMI + 10% FBS containing 6 × 106 differentiated HL-60 cells was added to the wells, and the plate was incubated for 90 min at 37°C with 5% CO2. Wells were aspirated, and the supernatant was diluted between 1:200,000 and 1:400,000 and plated (50 μl) onto LB agar. Two hundred microliters of 2 μM PelAh and 2 μM PslGh was added to aspirated wells, and the plate was incubated at room temperature on the nutator for 1 to 1.5 hours. The wells were aspirated, diluted, and plated onto LB agar as described above.
Cell morphology and viability assays
IMR-90 human lung fibroblast cells were seeded in 96-well CellBIND plates (Corning) at a density of 8000 to 10,000 cells per well. The next day, the medium was exchanged with a medium containing 1 μM CellTracker Orange CMRA (Molecular Probes) in serum-free Eagle’s minimum essential medium (Wisent) or supplemented with 10% FBS. After 60 min, excess dye was removed by media exchange. PslGh and PelAh were added to a final concentration of 1 mg/ml. The detergent digitonin and the C. difficile toxin TcdB were utilized as controls and added to a final concentration of 0.03 mg/ml and 0.5 pM, respectively (62). The cell plates were returned to the incubator for 5 hours before imaging. CellTracker-labeled cells were evaluated on a Cellomics ArrayScan VTI HCS reader (Thermo Scientific) using the target acquisition mode, a 10× objective, and a sample rate of 100 objects per well. Following a 24-hour incubation, cells in serum-free medium were supplemented with 10% FBS. Forty-eight hours after incubation, PrestoBlue reagent was added to the cells at a 1:10 reagent/media ratio and allowed to incubate for 5 hours. The microtiter plates were read in a SpectroMax M2 plate reader with λex (excitation wavelength) of 555 nm and λem (emission wavelength) of 585 nm. To probe the presence of PslGh and PelAh, we conducted Western blot analysis using polyclonal antibodies that were previously generated against both enzymes (47, 60).
Statistical analysis
One-way analysis of variance and Tukey’s multiple comparison test were utilized to evaluate statistical significance. EC50 values were calculated using nonlinear least-squares fitting to a dose-response model in Prism (GraphPad software). EC50 was the concentration of glycoside hydrolase required to reduce the biofilm biomass by half after a specified treatment time. All means, SEM, bar graphs, and dose-response curves were calculated and generated using Prism 6.0h.
Acknowledgments
Funding: This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR) (grant 43998 to P.L.H., grant 123306 to D.C.S., grant 286650 to R.A.M., and grant 81361 to P.L.H. and D.C.S.), Cystic Fibrosis Canada (CFC) (to D.C.S. and P.L.H.), the NIH (grant R01AI097511 to D.J.W. and grant 2R01AI077628 to M.R.P.), and the Natural Sciences and Engineering Research Council of Canada (grant RGPIN 418405 to R.A.M.). P.B. has been supported in part by a CFC postdoctoral fellowship and a Banting Fellowship from CIHR. B.D.S. has been supported by graduate scholarships from CFC and CIHR. L.K.J. is the recipient of an American Heart Association Postdoctoral Fellowship (14POST20130017). P.L.H. is the recipient of a Canada Research Chair. Author contributions: P.B., M.R.P., D.C.S., D.J.W., and P.L.H. designed the study and wrote the paper. P.B. completed the biofilm inhibition, biofilm disruption, antibiotic, and cytotoxicity assays. P.J.H. and M.J.P. completed the biofilm disruption assays and confocal microscopy. B.D.S. and M.J.L. completed the neutrophil assays. N.A. designed the vectors for the expression and purification of the PelA hydrolase and performed the initial Pel biofilm inhibition studies. L.K.J. completed the confocal microscopy experiments on the Pel-dependent biofilms. P.B., J.T., and R.A.M. developed and completed the cytotoxicity experiments. All authors approved the final version of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/5/e1501632/DC1
table S1. Strains and plasmids used in this study.
fig. S1. Time course disruption of P. aeruginosa biofilms.
fig. S2. P. aeruginosa biofilm disruption by glycoside hydrolases in the presence of serum.
fig. S3. Biofilm prevention in standing culture pellicle assay.
fig. S4. Protein stability of PelAh and PslGh in P. aeruginosa culture.
fig. S5. The growth of P. aeruginosa in the presence of glycoside hydrolases.
fig. S6. Protein stability of PelAh and PslGh in mammalian cell culture.
REFERENCES AND NOTES
Articles from Science Advances are provided here courtesy of American Association for the Advancement of Science
Full text links
Read article at publisher's site: https://doi.org/10.1126/sciadv.1501632
Read article for free, from open access legal sources, via Unpaywall:
https://advances.sciencemag.org/content/advances/2/5/e1501632.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The role of exopolysaccharides Psl and Pel in resistance of <i>Pseudomonas aeruginosa</i> to the oxidative stressors sodium hypochlorite and hydrogen peroxide.
Microbiol Spectr, 12(10):e0092224, 28 Aug 2024
Cited by: 0 articles | PMID: 39194290 | PMCID: PMC11448232
Molecular Mechanisms of Biofilm Formation in <i>Helicobacter pylori</i>.
Antibiotics (Basel), 13(10):976, 16 Oct 2024
Cited by: 0 articles | PMID: 39452242 | PMCID: PMC11504965
Review Free full text in Europe PMC
Resin Acid Copper Salt, an Interesting Chemical Pesticide, Controls Rice Bacterial Leaf Blight by Regulating Bacterial Biofilm, Motility, and Extracellular Enzymes.
Molecules, 29(18):4297, 11 Sep 2024
Cited by: 0 articles | PMID: 39339292 | PMCID: PMC11434517
An exopolysaccharide pathway from a freshwater Sphingomonas isolate.
J Bacteriol, 206(8):e0016924, 15 Jul 2024
Cited by: 0 articles | PMID: 39007563 | PMCID: PMC11340318
Exploring marine-derived bioactive compounds for dual inhibition of Pseudomonas aeruginosa LpxA and LpxD: integrated bioinformatics and cheminformatics approaches.
Mol Divers, 23 May 2024
Cited by: 0 articles | PMID: 38780832
Go to all (126) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AAG06452
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Treatment with the Pseudomonas aeruginosa Glycoside Hydrolase PslG Combats Wound Infection by Improving Antibiotic Efficacy and Host Innate Immune Activity.
Antimicrob Agents Chemother, 63(6):e00234-19, 24 May 2019
Cited by: 47 articles | PMID: 30988141 | PMCID: PMC6535529
Microbial glycoside hydrolases as antibiofilm agents with cross-kingdom activity.
Proc Natl Acad Sci U S A, 114(27):7124-7129, 20 Jun 2017
Cited by: 55 articles | PMID: 28634301 | PMCID: PMC5502622
Non-eluting, surface-bound enzymes disrupt surface attachment of bacteria by continuous biofilm polysaccharide degradation.
Biomaterials, 167:168-176, 14 Mar 2018
Cited by: 14 articles | PMID: 29571052
Regulation of Biofilm Exopolysaccharide Biosynthesis and Degradation in Pseudomonas aeruginosa.
Annu Rev Microbiol, 76:413-433, 02 Jun 2022
Cited by: 27 articles | PMID: 35655342
Review
Funding
Funders who supported this work.
Canadian Institutes of Health Research (8)
Grant ID: ID0EBOAK7609
Grant ID: 286650
Grant ID: ID0EJBAK7606
Grant ID: ID0E5JAK7608
Grant ID: 43998
Grant ID: 123306
Grant ID: 81361
Grant ID: ID0E2FAK7607
Cystic Fibrosis Canada (1)
Grant ID: ID0EESAK7610
NIAID NIH HHS (2)
Grant ID: R01 AI077628
Grant ID: R01 AI097511
National Institutes of Health (4)
Grant ID: 2R01AI077628
Grant ID: ID0EX2AK7612
Grant ID: R01AI097511
Grant ID: ID0EFWAK7611
Natural Sciences and Engineering Research Council of Canada (2)
Grant ID: ID0EY6AK7613
Grant ID: 418405





